In this post, we demonstrate how to use generative AI and Amazon Bedrock to transform natural language questions into graph queries to run against a knowledge graph. We explore the generation of queries written in the SPARQL query language, a well-known language for querying a graph whose data is represented as Resource Description Framework (RDF). The problem of mapping a natural language question to a SPARQL query is known as the text-to-SPARQL problem. This problem is similar to the much-studied text-to-SQL problem. We tackle this problem in a specific life-sciences domain: proteins.
Our approach allows a scientific user, knowledgeable of proteins but not of graph queries, to ask questions about proteins. We use a large language model (LLM), that we call via Bedrock, to generate from the user’s natural-language question a SPARQL query. We prompt the LLM to create a query that can be run against the established public UniProt dataset, an RDF dataset that can be queried with SPARQL. The LLM doesn’t actually run that query. We run it against a graph database containing UniProtKB. One such database is Amazon Neptune, a managed graph database service. Another is UniProt’s own public SPARQL endpoint.
UniProt Knowledgebase (UniProtKB), a non-trivial production knowledge base, is a worthy test. We show that a combination of few-shot prompting, natural language instructions, and an agentic workflow is efficacious at generating accurate SPARQL queries against UniprotKB.
The source for everything discussed in this post can be found in the accompanying GitHub repo. Please note that you will be responsible for the costs of the AWS resources used in the solution. A detailed breakdown of the estimated costs can be found in the GitHub repo here.
UniProtKB
The UniProtKB is a central resource for protein functional information, providing accurate and consistent annotations. It includes essential data for each protein entry—such as amino acid sequences, protein names, taxonomy, and references—along with comprehensive annotations, classifications, cross-references, and quality indicators based on experimental and computational evidence.
The UniProtKB is interesting for our purposes because it isn’t a test schema: it’s a large and complicated knowledge base that is used in many life sciences companies.
Amazon Neptune
Amazon Neptune is a managed graph database service from AWS designed to work with highly connected datasets, offering fast and reliable performance. Besides RDF and SPARQL, Neptune also supports labeled property graph (LPG) representation that can be queried using the Gremlin and openCypher query languages.
Our Git repo walks you through the steps to load UniProtKB data into your own Neptune database cluster. Neptune is optional in our design. You can walk through our example using the public UniProt SPARQL endpoint instead of using Neptune. UniProtKB’s size is large, on the order of hundreds of GB. As Exploring the UniProt protein knowledgebase with AWS Open Data and Amazon Neptune explains, that data can take some time to load into a Neptune database.
The advantage of having the data in Neptune is that you can then enrich it, analyze it, and combine it with other scientific data. But if you prefer a quick-start, you may skip the Neptune load and use the UniProt SPARQL endpoint instead.
Amazon Bedrock
Amazon Bedrock is a managed service that allows businesses to build and scale generative AI applications using foundation models (FMs) from top providers like AI21 Labs, Anthropic, and Stability AI, as well as Amazon’s own models, such as Amazon Nova. Amazon Bedrock provides access to a variety of model types, including text generation, image generation, and specialized models, allowing you to select the best fit for your use case. With Amazon Bedrock, developers can customize these models without needing extensive machine learning (ML) expertise, using their proprietary data securely for tailored applications. This service integrates with other AWS tools, such as Amazon SageMaker, facilitating the end-to-end development and deployment of AI applications across diverse industries.
Solution overview
There has been a lot of successful work on the text-to-SQL problem (converting natural language questions to SQL queries), and it’s natural to assume that those same techniques would map readily to the text-to-SPARQL problem. In particular, solutions to the text-to-SQL problem rely heavily on providing the model with an explicit representation of the relational schema (typically a subset of the DDL or CREATE TABLE statements). We found experimentally that giving the model an explicit representation of the schema did not help the model generate accurate SPARQL queries.
For the text-to-SPARQL problem, rather than using an explicit representation of the schema, we use the following design patterns:
- We provide in the prompt a set of few-shot examples of known pairs of questions and SPARQL queries
- We provide a set of natural language descriptions of parts of the schema
- We use an agentic workflow to allow the model to critique and correct its own work
We tested this approach with Anthropic’s Claude 3.5 Sonnet v2 on Amazon Bedrock. Less powerful models are cheaper and faster, but not as good at following instructions and will not fully use our set of natural language instructions.
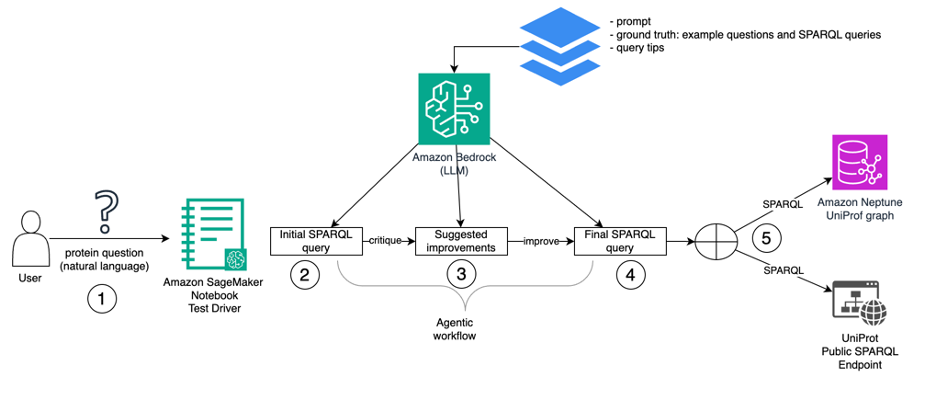
The follow diagram illustrates the system architecture.

Our test driver is a Jupyter notebook instance running on Amazon SageMaker. The sequence of steps from natural language to SPARQL query execution is the following:
- The user’s natural-language question is taken as input.
- The driver begins the first step of its agentic workflow, calling the LLM via Bedrock to generate an initial SPARQL query based on the natural-language question. The driver provides a prompt, as well as a set of ground truth examples (typical natural language questions about proteins and their corresponding SPARQL queries) and tips (conventions to use while forming SPARQL queries).
- The driver prompts the LLM to critique its initial query. The output is a set of improvements.
- The driver prompts the LLM to generate a final SPARQL query based on its critique.
- The driver executes the final SPARQL query against the graph database. It can run it either against your Neptune database cluster or the public UniProt SPARQL endpoint.
Few-shot examples
We compiled a collection of 44 question-and-SPARQL-query pairs from the UniProt website. The following is one example pair:
question: Select reviewed UniProt entries (Swiss-Prot), and their recommended
protein name, that have a preferred gene name that contains the text ‘DNA’
SPARQL:
We didn’t include all of the SPARQL examples that are on the UniProt website; we excluded those that construct new triples and those that use external services.
We use a subset of the resulting 44 question-and-SPARQL-query pairs as few-shot examples. When testing a question, we do a form of leave-one-out validation; we test on one of the known 44 questions and include the other 43 question-SPARQL pairs as few-shot examples in the prompt.
Natural language instructions
We use natural language statements (hints) to help the language model understand the UniProt schema. The following are some examples:
- To find the name of a GO class use
rdfs:label - The category of an external database is encoded as
up:categorywhere the object is the name of the category - The IRIs for annotations come in two forms. The first is like this,
<http://purl.uniprot.org/uniprot/P83549#SIP0DB7D53171472E13>, the accession number is the last part of the IRI:P83549#SIP0DB7D53171472E13. The second is like this,<http://purl.uniprot.org/annotation/VAR_000007>, and the accession number here is again the last part of the IRI:VAR_000007.
Each hint tells the model about one aspect of the UniProt schema. We currently have 59 such hints and Anthropic’s Claude 3.5 Sonnet v2 will follow all of these hints. Note that less powerful models won’t necessarily follow all of the hints.
Agentic workflow
An agentic workflow is a connected set of model inferences that collectively solves a problem. In our case, we use separate inferences to create a SPARQL query, the model then critiques that query, and then the model uses that critique to modify the query. This allows us to distribute the computation over multiple inferences, allowing the total computation to be more powerful and accurate than what could be achieved by a single inference.
In this particular case, our agentic workflow has three steps and it consistently performs the same three steps in this order:
- Given the natural language question, hints, and few-shot examples, the model generates an initial SPARQL query.
- Given the initial query, question, and hints, the model critiques this query and returns a list of suggested changes that would improve the query (either in terms of accuracy or speed).
- Given the initial query and a list of suggestions, the model implements those changes and returns the updated final query.
Results
We tested our approach on the preceding set of 44 question-SPARQL pairs. Of these, 61% were correct, 23% were wrong, and for 16% we were unsure (the ground truth and generated queries were too complicated). To help understand when this approach does and doesn’t work, we can closely examine two examples, one that was successful and one that wasn’t.
A successful example
For the question, “Select all human UniProt entries with a sequence variant that leads to a tyrosine to phenylalanine substitution,” our system generated the following SPARQL query:
We can see here that the system has learned at least the following about the UniProt schema:
- The appropriate use of the properties
up:organism,up:annotation,up:substitution, and so on - How to use the
faldo:ontology - How the UniProt ontology encodes the variant and original sequences
- A (limited) understanding of the
taxon:hierarchy
An unsuccessful example
For the question, “Select the number of UniProt entries for each of the EC (Enzyme Commission) second level categories,” the ground truth SPARQL query is as follows:
Whereas our system generates the following incorrect query:
Our system appears unable to understand how to generate queries that involve terms like ec:1._._._ and so such queries are beyond the current ability of our text-to-SPARQL system.
Clean up
To avoid future charges, our Git repo shows how to delete the AWS resources you created.
Conclusion
In this post, we showed that an agentic architecture along with few-shot prompting and natural language instructions (hints) can achieve an accuracy of 61.4% on our test suite.
An independent study, GenAI Benchmark II: Increased LLM Accuracy with Ontology-Based Query Checks and LLM Repair, adds evidence to our hypothesis that an agentic architecture can improve the performance of SPARQL generation.
We encourage you to take this work and extend it to your own knowledge base. To do this, you should start with modifying the following files in our GitHub repo:
- tips.yaml contains the hints to the model that should be replaced with hints that are specific to your particular ontology.
- ground-truth.yaml contains the few-shot examples. Replace our examples with examples that are specific to your ontology.
- prefixes.txt contains the prefixes that are used by your ontology.
Once these files are updated to correspond to your ontology, you can use the run_gen_tests.ipynb Jupyter notebook to generate SPARQL queries.
About the authors
 Simon Handley, PhD, is a Senior AI/ML Solutions Architect in the Global Healthcare and Life Sciences team at Amazon Web Services. He has more than 25 years’ experience in biotechnology and machine learning, and is passionate about helping customers solve their machine learning and genomic challenges. In his spare time, he enjoys horseback riding and playing ice hockey.
Simon Handley, PhD, is a Senior AI/ML Solutions Architect in the Global Healthcare and Life Sciences team at Amazon Web Services. He has more than 25 years’ experience in biotechnology and machine learning, and is passionate about helping customers solve their machine learning and genomic challenges. In his spare time, he enjoys horseback riding and playing ice hockey.
 Mike Havey is a software architect with 30 years of experience building enterprise applications. Mike is the author of two books and numerous articles. Visit his Amazon author page.
Mike Havey is a software architect with 30 years of experience building enterprise applications. Mike is the author of two books and numerous articles. Visit his Amazon author page.
Source: Read More