Enzymes are indispensable molecular catalysts that facilitate the biochemical processes vital to life. They play crucial roles across metabolism, industry, and biotechnology. Despite their importance, there are significant gaps in our knowledge of these catalysts. Out of the approximately 190 million protein sequences cataloged in databases like UniProt, fewer than 0.3% are curated by experts, and less than 20% have experimental validation. Furthermore, 40-50% of known enzymatic reactions remain unlinked to specific enzymes, often termed “orphaned†reactions. These knowledge gaps hinder progress in synthetic biology and biotechnological innovation. Traditional computational tools, including EC classification and sequence-similarity methods, frequently fall short, particularly when dealing with enzymes of low sequence homology or reactions that do not align with established classifications. To overcome these limitations, new strategies that combine structural and functional insights are needed.
EnzymeCAGE: A New Approach
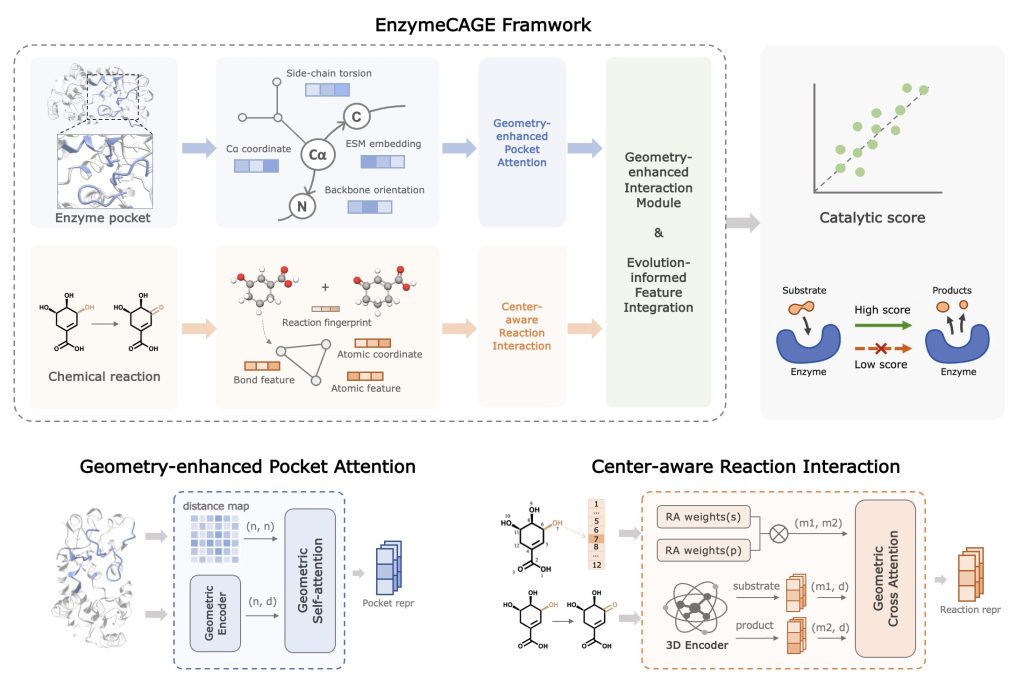
A team of researchers from Shanghai Jiaotong University, Hong Kong University of Science and Technology, Hainan University, Sun Yat-sen University, McGill University, Mila-Quebec AI Institute, and MIT developed a new open-sourced foundation model for enzyme retrieval and function prediction called EnzymeCAGE. This model is trained on a dataset of approximately one million enzyme-reaction pairs and employs the Contrastive Language–Image Pretraining (CLIP) framework to annotate unseen enzymes and orphan reactions. EnzymeCAGE, an acronym for CAtalytic-aware GEometric-enhanced enzyme retrieval model, integrates structural learning with evolutionary insights to address the limitations of conventional methods. The model effectively links unannotated proteins with catalytic reactions and identifies enzymes for novel reactions. EnzymeCAGE is a robust tool for enzymology and synthetic biology by leveraging enzyme structures and reaction mechanisms. It’s geometry-aware and reaction-guided modules allow for precise insights into enzyme catalysis, making it applicable to a wide range of species and metabolic contexts.

Technical Features and Benefits
EnzymeCAGE incorporates several advanced features to model enzyme-reaction interactions effectively. At its core is the geometry-enhanced pocket attention module, which utilizes structural information such as residue distances and dihedral angles to pinpoint catalytic sites. This enhances both the accuracy and interpretability of its predictions. Additionally, the model employs a center-aware reaction interaction module that emphasizes reaction centers through weighted attention, capturing the dynamics of substrate-product transformations. EnzymeCAGE combines local pocket-level encoding using Graph Neural Networks (GNNs) with global enzyme-level features from the ESM2 protein language model. This holistic approach provides a comprehensive representation of catalytic potential. Furthermore, the model’s compatibility with both experimental and predicted enzyme structures broadens its applicability to tasks such as enzyme retrieval, reaction de-orphaning, and pathway engineering.
Performance and Insights
EnzymeCAGE has undergone rigorous testing, demonstrating superior performance compared to existing methods. In the Loyal-1968 test set, which featured unseen enzymes, the model achieved a 44% improvement in function prediction and a 73% increase in enzyme retrieval accuracy relative to traditional approaches. It recorded a Top-1 success rate of 33.7% and a Top-10 success rate exceeding 63%, outperforming benchmarks like BLASTp and Selenzyme. In reaction de-orphaning tasks, EnzymeCAGE consistently identified suitable enzymes for orphan reactions, achieving higher enrichment factors and ranking metrics across diverse test sets. Practical case studies further highlight its capabilities, including the accurate reconstruction of the glutarate biosynthesis pathway, where it surpassed traditional methods in ranking and selecting enzymes. These results underscore EnzymeCAGE’s utility in tackling major challenges in enzyme function prediction and catalysis research.
Conclusion
EnzymeCAGE represents a significant step forward in addressing longstanding challenges in enzyme research, particularly in function prediction and reaction annotation. By integrating geometric, structural, and functional insights, it delivers accurate predictions for unseen enzyme functions, annotations for orphan reactions, and support for pathway engineering. The model’s adaptability and fine-tuning capabilities enhance its utility for specific enzyme families and industrial applications. EnzymeCAGE sets a strong foundation for future advancements in biocatalysis, synthetic biology, and metabolic engineering, offering new avenues to deepen our understanding of enzymatic processes and their potential for innovation.
Check out the Paper and GitHub Page. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. Don’t Forget to join our 60k+ ML SubReddit.
The post EnzymeCAGE: A Deep Learning Framework Designed to Predict Enzyme-Reaction Catalytic Specificity by Encoding both Pocket-Specific Enzyme Structures and Chemical Reactions appeared first on MarkTechPost.
Source: Read MoreÂ
 Trending: LG AI Research Releases EXAONE 3.5: Three Open-Source Bilingual Frontier AI-level Models Delivering Unmatched Instruction Following and Long Context Understanding for Global Leadership in Generative AI Excellence….
Trending: LG AI Research Releases EXAONE 3.5: Three Open-Source Bilingual Frontier AI-level Models Delivering Unmatched Instruction Following and Long Context Understanding for Global Leadership in Generative AI Excellence….