Trailing the advances made by AI in drug discovery, one can say there is a vast amount of untapped potential. Therapeutic nanobodies, particularly, have had relatively limited breakthroughs as they require complex interdisciplinary knowledge. The COVID-19 pandemic urged the development of therapeutic nanobodies that exhibit high binding affinity and stability for the SARS-CoV-2 in a short period. However, developing and testing a new drug is a resource-intensive and time-consuming. Researchers at the Department of Computer Science and Biomedical Data Science, Stanford University, and Chan Zuckerberg Biohub, San Francisco, have used a notable framework, Virtual Lab, that has helped streamline the drug development process from its designing to testing.Â
Conventional methods involve experimental screening of large libraries of nanobody candidates against the target antigen to identify high-affinity binders. However, it requires significant time, resources, and labor. Computational methods have also been developed to identify the nanobody candidates, but they have been found to lack accuracy, which could be very detrimental if used as a therapeutic. Given the rapid mutation rates of the SARS-CoV-2 virus, it is imperative that a substantial amount of lives will be lost while the drugs are in the process of development. These limitations have put a strain on the healthcare system.Â
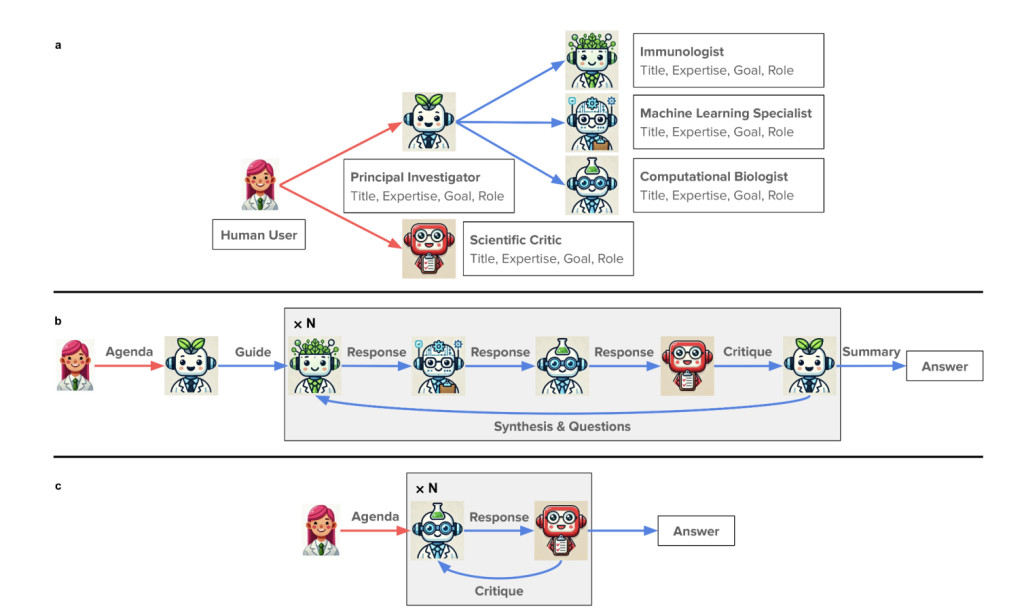
The proposed method employs a virtual lab environment where AI agents with different areas of expertise collaborate and tackle the problem, mimicking real-world scientific teamwork. A computational pipeline is developed after conducting meetings between the AI agents. The key components of this pipeline include:
- ESM (Evolutionary Scale Modeling): It analyses the protein sequences and notes the effects of various mutations on the protein function and stability. This tool is critical to finding potential mutations that enhance the nanobody binding to our virus’ spike proteins.Â
- AlphaFold-Multimer: To predict the protein-protein interaction between the virus and nanobody, AplhaFold-Multimer uses deep learning and generates high-confidence structural predictions.Â
- Rosetta: It uses the iterative refinement process to optimize the three-dimensional structures of the designed nanobodies.

Experimental validation showed that more than 90% of the engineered nanobodies were expressed and soluble, and two candidates displayed superior binding properties specifically against the new JN.1 and KP.3 variants of SARS-CoV-2 while retaining solid interactions with the ancestral spike protein. This is an essential result for demonstrating the effectiveness of the Virtual Lab’s computational framework in generating viable therapeutic candidates quickly.
In conclusion, this paper describes AI-based nanobodies produced with incorporation into the existing experimental methodologies. Such a synergistic framework of several artificial agents highly elevates the stages of design and validation from many established methods, which tend to be very time- and resource-consuming. Optimal identification of the directed nanobodies against the SARS-CoV-2 variants provides essential evidence that AI may prove critical in speeding up therapeutical discoveries. This novel approach enhances effectiveness in nanobody design and facilitates quick response to emergent viral threats. This gives it an outlook that outlines the tremendous effect of artificial intelligence in biomedical research and its applications in developing therapy.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter.. Don’t Forget to join our 59k+ ML SubReddit.
The post The Virtual Lab: AI Agents Design New SARS-CoV-2 Nanobodies with Experimental Validation appeared first on MarkTechPost.
Source: Read MoreÂ



 ‘
‘