Drug discovery is a costly, lengthy process with high failure rates, as only one viable drug typically emerges from a million screened compounds. Advanced high-throughput (HTS) and ultra-high-throughput screening (uHTS) technologies allow rapid testing of large compound libraries, enabling Pharma and Biotech companies to explore more chemical compounds and novel biological targets. Despite these technologies, challenges still need to be addressed, including limited breakthroughs in identifying new drug targets and data quality issues. ML and DL now offer promising solutions, enhancing drug discovery through data-driven insights, feature extraction, and predictive capabilities to identify effective drug candidates more efficiently.
VirtuDockDL, developed by researchers from the Institute of Molecular Biology and Biotechnology at The University of Lahore, the Integrative Omics and Molecular Modeling Laboratory at Government College University Faisalabad (GCUF), Shenzhen University and Taif University, is a Python-based platform leveraging deep learning to streamline drug discovery. Utilizing a Graph Neural Network (GNN) for predicting compound effectiveness, VirtuDockDL achieved 99% accuracy on the HER2 dataset, surpassing tools like DeepChem and AutoDock Vina. This platform’s automated framework integrates molecular graph construction, virtual screening, and compound clustering, enabling efficient identification of potential drugs and advancing AI-driven pharmaceutical research.
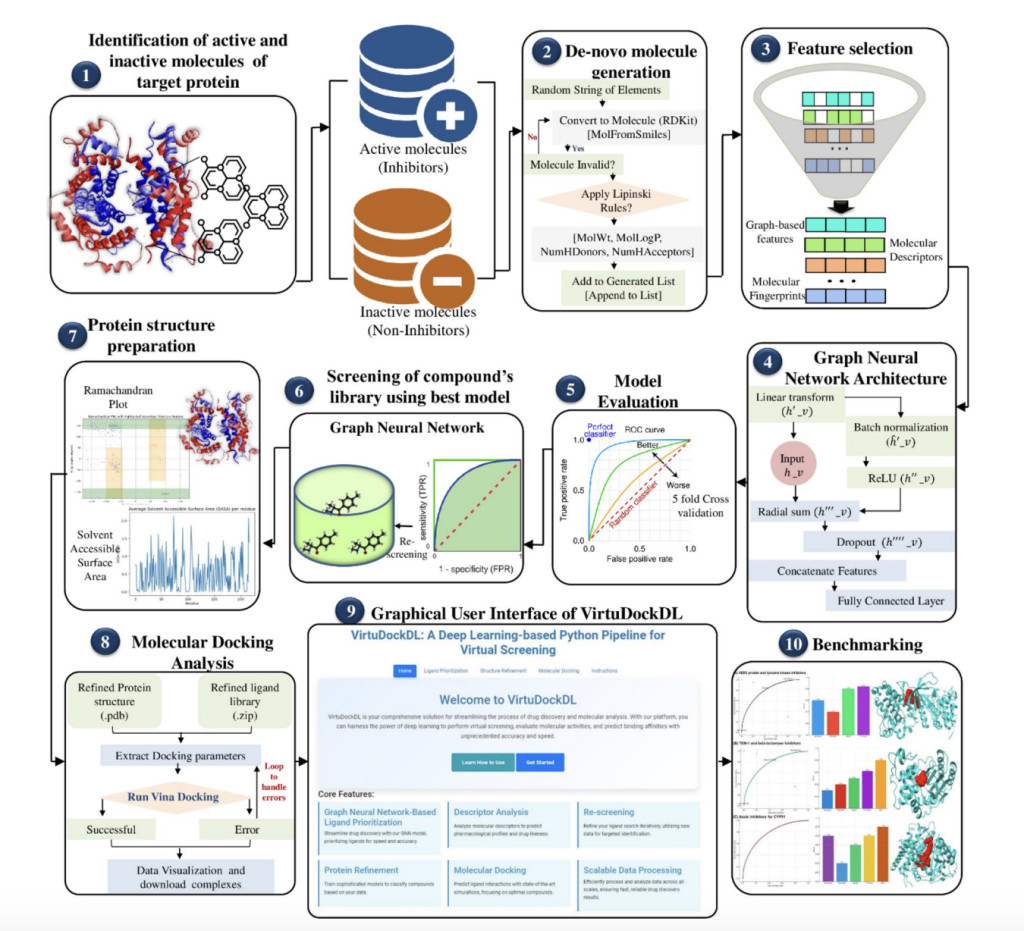
VirtuDockDL is a comprehensive pipeline designed to streamline the prediction and screening of biologically active compounds using a GNN. Initially encoded as SMILES strings, molecular data is transformed into graph representations through RDKit and processed by PyTorch Geometric’s GNN architecture. This transformation allows the GNN to learn complex structural relationships within molecules and predict properties like molecular activity or binding affinity. The architecture incorporates several layers of graph convolution to capture molecular features at different hierarchical levels, along with batch normalization, dropout, and residual connections, which stabilize training and enhance predictive accuracy. This process merges graph-based representations with cheminformatics descriptors and fingerprints, providing a robust feature set for accurate activity prediction.
The application also features virtual screening and clustering tools, enabling users to evaluate large compound libraries against specific protein targets. Based on their predicted activity, the clustering of screened molecules is accomplished using Gaussian Mixture Models (GMM), with clustering quality assessed via Silhouette and Davies-Bouldin scores. The pipeline supports protein structure refinement through OpenMM and ligand docking with AutoDock Vina, allowing molecular binding affinity predictions. VirtuDockDL was applied to Marburg virus research, using the VP35 protein as a case study. Positive and decoy datasets were generated, and the GNN model accurately classified compounds with cross-entropy loss and RMSprop optimization. Virtual screening and docking results, including key metrics like AUC, accuracy, and F1-score, are automatically visualized, providing actionable insights into potential VP35 inhibitors for drug discovery.
VirtuDockDL’s user-friendly GUI, based on the Flask framework, supports molecule uploads, task initiation, and result downloads, organizing features into tabs for ease of use. A GNN model was trained using active/inactive VP35 protein molecules, achieving high accuracy (97.79%) with strong metrics (AUC 0.9972). Non-covalent inhibitors from ZINC and PubChem databases were re-screened, identifying 146 potential candidates. Further tests on HER2, beta-lactamase, and CYP51 datasets demonstrated VirtuDockDL’s superior performance in binding affinity predictions compared to PyRMD, RosettaVS, MzDOCK, AutoDock Vina, and Glide. VirtuDockDL’s integration of ligand- and structure-based screening provides efficient and accurate virtual screening.
In conclusion, VirtuDockDL is a new Python-based web platform designed to streamline drug discovery using deep learning. By employing a Graph Neural Network for compound screening, it has shown outstanding predictive accuracy and practical utility across multiple targets, including inhibitors for HER2 (cancer), TEM-1 beta-lactamase (bacterial infections), and CYP51 (Candidiasis). It achieved superior results in benchmarking, surpassing tools like DeepChem and AutoDock Vina with a 99% accuracy and an F1 score of 0.992 on the HER2 dataset. This platform combines full automation and user-friendly design, making it an efficient, cost-effective tool for advancing pharmaceutical research and addressing urgent health challenges.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter.. Don’t Forget to join our 55k+ ML SubReddit.
[FREE AI WEBINAR] Implementing Intelligent Document Processing with GenAI in Financial Services and Real Estate Transactions– From Framework to Production
The post VirtuDockDL: A Deep Learning-Powered Platform for Accelerated Drug Discovery through Advanced Compound Screening and Binding Prediction appeared first on MarkTechPost.
Source: Read MoreÂ