Microscopic imaging is crucial in modern medicine as an indispensable tool for researchers and clinicians. This imaging technology allows detailed examination of biological structures at the cellular and molecular levels, enabling the study of tissue samples in disease diagnosis and pathology. By capturing these microscopic images, medical professionals can better understand disease mechanisms and progression, often revealing subtle changes not detectable through other methods. However, despite the importance of these images, their classification and interpretation usually demand specialized expertise and substantial time investment, leading to inefficiencies in diagnosis. As the volume of medical data grows, the demand for automated, efficient, and accurate tools for microscopic image classification has become more pressing.
A key issue in medical image classification is the challenge of effectively interpreting and classifying these complex images. Manual classification is slow and prone to inconsistencies due to the subjective nature of human judgment. Moreover, the scale of the data generated through microscopic imaging makes manual analysis impractical in many scenarios. Traditional machine learning methods, such as convolutional neural networks (CNNs), have been employed for this task, but they come with limitations. While CNNs are powerful in extracting local features, their ability to capture long-range dependencies across the image is limited. This restriction prevents them from fully utilizing the semantic information embedded in medical images, which is critical for accurate classification and diagnosis. On the other hand, vision transformers (ViTs), known for their efficiency in modeling global dependencies, suffer from high computational complexity, particularly in long-sequence modeling, which renders them less suitable for real-time medical applications where computational resources may be limited.
Existing methods to address these limitations have included hybrid approaches combining CNNs and transformers. These methods attempt to balance between local and global feature extraction but often come at the cost of either accuracy or computational efficiency. Some studies have proposed reduced-complexity ViTs to make them more feasible for practical use. However, these models often sacrifice precision in medical imaging, where every pixel’s information could be crucial for accurate diagnosis. Thus, there is a clear need for more efficient models that can effectively handle both local and global information without a significant computational burden.
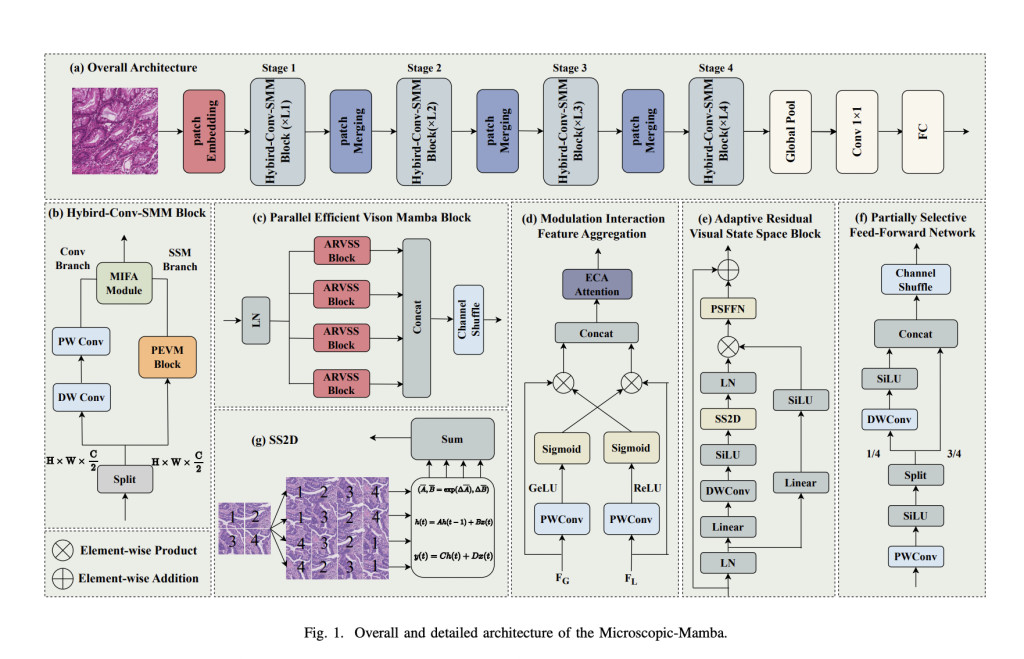
A research team from Nanjing Agricultural University, National University of Defense Technology, Xiangtan University, Nanjing University of Posts and Telecommunications, and Soochow University introduced a novel architecture called Microscopic-Mamba to address these challenges. This hybrid model was specifically designed to improve microscopic image classification by combining the strengths of CNNs in local feature extraction with the efficiency of State Space Models (SSMs) in capturing long-range dependencies. The team’s model integrates the Partially Selected Feed-Forward Network (PSFFN) to replace the final linear layer in the Vision State Space Module (VSSM), significantly enhancing the ability to perceive local features while maintaining a compact and efficient architecture. By incorporating global and local information processing capabilities, the Microscopic-Mamba model seeks to set a new benchmark in medical image classification.
The core methodology behind Microscopic-Mamba lies in its dual-branch structure, consisting of a convolutional branch for local feature extraction and an SSM branch for global feature modeling. The model also introduces the Modulation Interaction Feature Aggregation (MIFA) module, designed to effectively fuse global and local features. In this architecture, the CNN branch uses depth-wise separable convolution (DWConv) and point-wise convolution (PWConv) for localized feature extraction. In contrast, the SSM branch focuses on global feature modeling through the parallel Vision State Space Module (VSSM). Integrating these two modules allows Microscopic-Mamba to process detailed local information and broad global patterns, which is essential for accurate medical image analysis. The final layer in the VSSM is replaced with the PSFFN, which refines the model’s ability to capture local information, optimizing the balance between detail and generalization.
The Microscopic-Mamba model demonstrated superior performance on five public medical image datasets in extensive testing. These datasets included the Retinal Pigment Epithelium (RPE) Cell dataset, the SARS dataset for malaria cell classification, the MHIST dataset for colorectal polyp classification, the MedFM Colon dataset for tumor tissue classification, and the TissueMNIST dataset, which contains over 236,386 images of human kidney cells. The model achieved a remarkable balance of high accuracy and low computational demands, making it ideal for real-world medical applications. On the RPE dataset, for example, Microscopic-Mamba achieved an overall accuracy (OA) of 87.60% and an area under the curve (AUC) of 98.28%, outperforming existing methods. The model’s lightweight design, with only 4.49 GMACs and 1.10 million parameters on some tasks, ensures that it can be deployed in environments with limited computational resources while maintaining high accuracy.
Ablation studies showed that introducing the MIFA module and the PSFFN was critical to the model’s success. Combining these two elements led to notable improvements in performance across all datasets. On the MHIST dataset, the model achieved an AUC of 99.56% with only 4.86 million parameters, underscoring its efficiency and effectiveness in medical image classification.
In conclusion, the Microscopic-Mamba model significantly advances medical image classification. By combining the strengths of CNNs and SSMs, this hybrid architecture successfully addresses the limitations of previous methods, offering a solution that is both computationally efficient and highly accurate. The model’s ability to process and integrate local and global features makes it well-suited for microscopic image analysis. With its impressive performance across multiple datasets, Microscopic-Mamba has the potential to become a standard tool in automated medical diagnostics, streamlining the process and improving the accuracy of disease identification.
Check out the Paper and GitHub Page. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter..
Don’t Forget to join our 50k+ ML SubReddit
The post Microscopic-Mamba Released: A Groundbreaking Hybrid Model Combining Convolutional Neural Network CNNs and SSMs for Efficient and Accurate Medical Microscopic Image Classification appeared first on MarkTechPost.
Source: Read MoreÂ