Machine Learning Models for Predicting Prime Editing Efficiency:
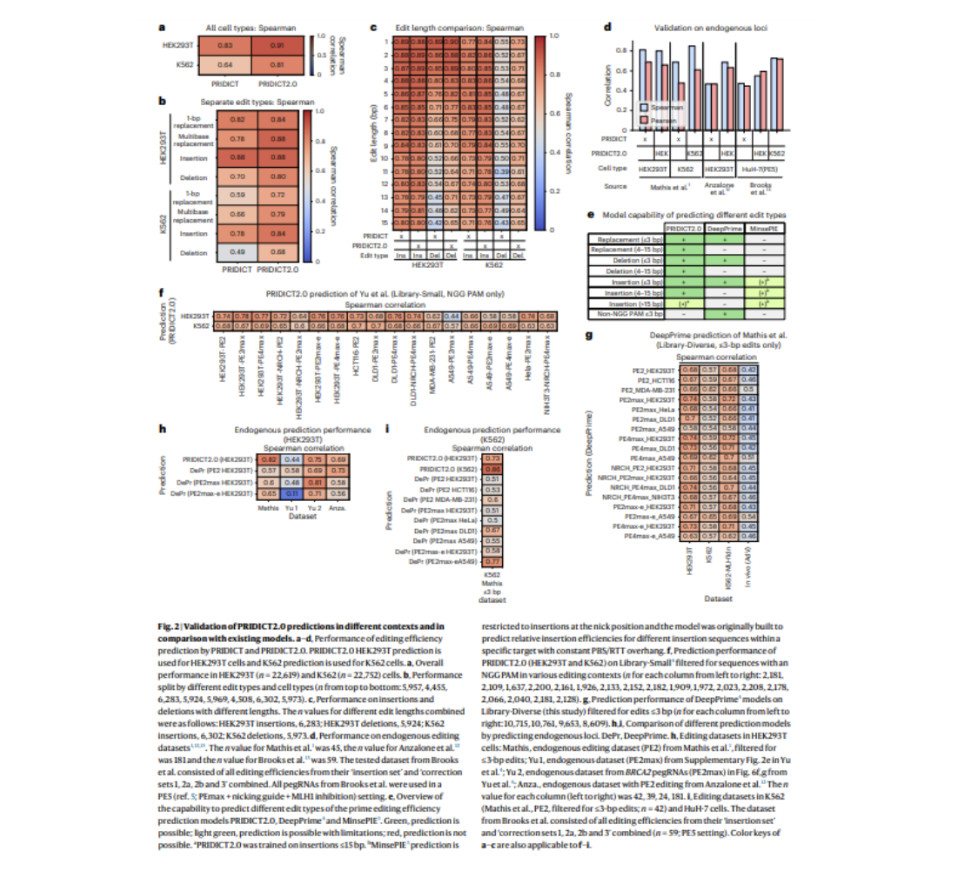
The success of prime editing is highly dependent on both the prime editing guide RNA (pegRNA) design and the target locus. To address this, researchers developed two complementary machine learning models—PRIDICT2.0 and ePRIDICT—to predict prime editing efficiency across various edit types and chromatin contexts. PRIDICT2.0, an advanced version of the earlier PRIDICT1 model, assesses pegRNA performance for edits up to 15 base pairs (bp) in mismatch repair (MMR)-deficient and proficient cell lines. At the same time, ePRIDICT quantifies how local chromatin environments impact prime editing rates. By utilizing a diverse pegRNA library in both HEK293T (MMR-deficient) and K562 (MMR-proficient) cells, the study demonstrated that PRIDICT2.0 significantly outperforms its predecessor, especially for multibase replacements and deletions. The model’s robustness was confirmed through extensive validation, showing strong correlations between experimental replicates and improved performance over previous models.
Insights into Chromatin Context and Editing Efficiency:
One of the key advancements in this study is the inclusion of chromatin context as a factor influencing prime editing efficiency. ePRIDICT was designed to predict editing outcomes by accounting for locus-specific chromatin features, adding a new layer of precision to editing predictions. Shapley additive explanations (SHAP) analysis revealed that features such as edit length, presence of polyT sequences, and RTT overhang length were highly relevant in HEK293T cells, while position, melting temperature, and G+C content played crucial roles in K562 cells. The study also found that editing patterns in MMR-deficient cells resembled those of K562 cells with suppressed MMR pathways, further reinforcing the importance of considering chromatin context for accurate predictions. Through these findings, the models offer valuable tools for improving pegRNA design and maximizing the efficiency of prime editing in diverse biological contexts.
Chromatin’s Role in Genome Editing Efficiency:
To explore chromatin’s influence on genome editing, cells were treated with ABE8e, BE4max, and Cas9, showing strong correlations in editing efficiency, especially between ABE8e and BE4max. Active chromatin features, like ATAC-seq and H3K4me3, positively correlated with editing efficiency, while repressive marks (H3K9me3, H3K27me3) were linked to lower efficiency. UMAP analysis highlighted a chromatin gradient influencing editing. The XGBoost-based ‘ePRIDICT’ model, trained on chromatin data, effectively predicted editing outcomes. Combining it with PRIDICT2.0 improved accuracy, especially in regions with lower chromatin accessibility, confirming chromatin’s pivotal role in editing outcomes.
Cloning and PegRNA Design:
The TRIP plasmid library used in chromatin-context studies was engineered according to a specified protocol. For validating the pegRNAs on endogenous targets, 20 genomic sites were selected from a prior screen—10 sites with high and 10 with low editing efficiency. PegRNAs were designed to achieve various genetic modifications: 1-bp replacements, 4-bp insertions, and 4-bp deletions. PegRNAs were selected based on their predicted editing efficiency and specific nucleotide presence within their target windows. In addition, 90 more pegRNAs targeting intronic and intergenic regions were designed and cloned using a particular vector. The sgRNAs were introduced into a plasmid via a one-pot cloning reaction, then transformed into competent bacterial cells, plasmid extraction, and verification.
Viral Vector Production and Screening:
Lentiviral and pseudotyped AAV9 vectors were produced by transfecting HEK293T cells with necessary plasmids and purifying the vectors through a series of precipitation and centrifugation steps. A separate viral vector containing a prime editing component was also produced. The pegRNA library, designed to include pathogenic variants and noncoding mutations, was ordered from a commercial provider. Various cell lines, including HEK293T, HepG2, and K562 cells, were maintained under specific conditions and subjected to transfection or electroporation for editing. The screening involved transducing cells with lentivirus and selecting edited cells using antibiotics. For in vivo studies, vectors were injected into mice, which were then euthanized for hepatocyte isolation. Genomic DNA from these experiments was isolated and analyzed using high-throughput sequencing techniques.
Library and Editing Efficiency Analysis:
Sequencing reads were trimmed and filtered to ensure accuracy, removing ~34% of reads in HEK293T and K562 cells and ~60% in mouse hepatocytes. Editing efficiency was calculated by comparing read sequences to wild-type and edited sequences, adjusting for background frequencies. PegRNAs were validated using specific criteria and averaged across replicates, resulting in several datasets. For the TRIP library, tagmentation was followed by PCR amplification and sequencing. Editing efficiencies were analyzed with custom scripts and cross-referenced with chromatin data from ENCODE. Machine learning models, including PRIDICT2.0, were trained and validated using various datasets, with performance evaluated by cross-validation and feature importance analysis.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter..
Don’t Forget to join our 50k+ ML SubReddit
The post Advancements in Machine Learning Models and Chromatin Context for Optimizing Prime Editing Efficiency appeared first on MarkTechPost.
Source: Read MoreÂ
![Why developers needn’t fear CSS – with the King of CSS himself Kevin Powell [Podcast #154]](https://devstacktips.com/wp-content/uploads/2024/12/15498ad9-15f9-4dc3-98cc-8d7f07cec348-fXprvk-450x253.png)