To diagnose and cure cancer, pathologic examination of tissue is crucial. The digital versions of the old histological slides used for light microscopy are gradually replacing them with whole-slide images (WSIs). This allows computational pathology to transition from being used primarily as academic proof points to becoming routine tools in clinical practice. To aid in diagnosing, characterizing, and comprehending disease, computational pathology uses digital WSIs in conjunction with artificial intelligence (AI). The first artificial intelligence pathology system to receive FDA approval was introduced in 2021, after initial efforts centered on clinical decision support tools to improve existing workflows. Newer research, however, aims to decipher regular WSIs for previously unknown outcomes like prediction and therapy response because of the remarkable performance advances in computer vision, an area of artificial intelligence centered around images.
Building large-scale deep neural networks, often called foundation models, has been a key component in computer vision model performance advancements. A class of algorithms called self-supervised learning is employed to develop foundation models. These models do not need curated labels and are trained on massive datasets, orders of magnitude larger than any traditionally used for computational pathology. Embeddings, the data representations generated by foundation models, can be generalized to various prediction tasks. This starkly contrasts the existing diagnostic-specific computational pathology methods, which only use a subset of pathology images and will not be able to generalize well enough due to the wide range of variations in tissue morphology and laboratory preparations. For applications lacking data to build custom models, such as rare tumor types detection or less common diagnostic tasks like predicting specific genomic alterations, clinical outcomes, or therapeutic response, the value of generalization from big datasets becomes even more apparent. This model has the potential to be used for a wide range of important tasks, including clinically robust cancer prediction (both common and rare), subtyping, biomarker quantification, cellular instance, event counting, and therapeutic response prediction, provided it is trained with a large enough quantity of digital WSIs in the pathology domain.
As shown by scaling law studies, the performance of foundational models is highly dependent on the size of the dataset and the model itself. To train models with hundreds of millions to billions of parameters, modern natural image domain foundation models use datasets like ImageNet, JFT-300M, and LVD-142M, among others. Another example is vision transformers (ViTs). In spite of the difficulties in gathering pathology-specific large-scale datasets, some groundbreaking research has used datasets with 30,000 to 400,000 WSIs to train 28 million to 307 million parameter foundation models. This groundbreaking research is inspiring and paves the way for further advancements in computational pathology.
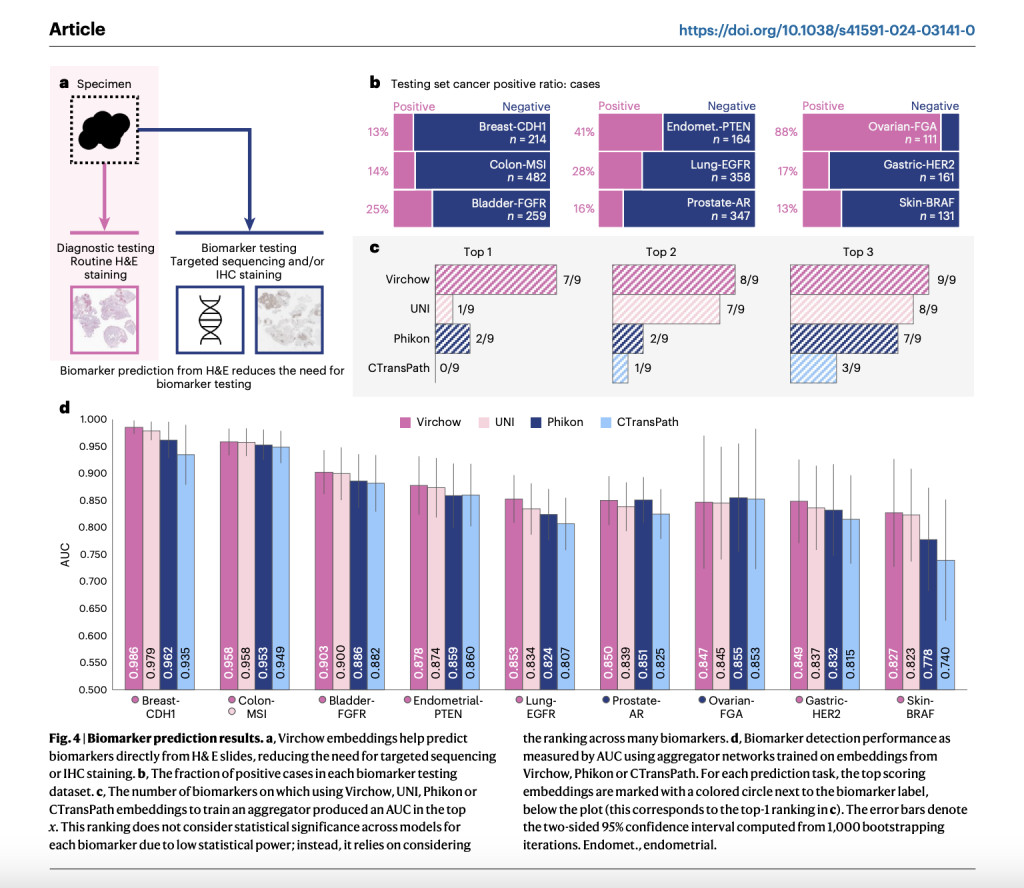
Virchow, a pathology foundation model with a million images, was developed by Paige and Microsoft Research. It is named after Rudolf Virchow, the man widely recognized as the father of modern pathology and who put forth the first theory of cellular pathology. Memorial Sloan Kettering Cancer Center (MSKCC) provided Virchow with 1.5 million H&E stained WSIs, representing around 100,000 patients. This is 4-10 times more WSIs than previous pathology teaching datasets. The training data set includes benign and malignant tissues derived from 17 different high-level tissues, with biopsy making up 63% and resection 37% of the total. Utilizing the DINO v. two algorithm—a multiview student-teacher self-supervised algorithm—Virchow, a ViT model with 632 million parameters, is trained. DINO v.2 uses global and local tissue tile areas to train various downstream prediction tasks to learn how to embed WSI tiles. These embeddings can then be pooled across slides.
The Virchow2 and 2G models are trained using the biggest known digital pathology dataset, which employs data from more than 3.1 million full slide pictures (2.4PB of data), representing more than 40 tissues from 225,000 patients in 45 countries. With 632 million parameters, Virchow2 is on par with the original Virchow model, and with 1.85 billion parameters, Virchow2G is the largest pathology model ever created. To back up this magnitude, researchers suggest domain-inspired modifications to the DINOv2 training algorithm—the de facto standard in computational pathology self-supervised learning. This algorithm plays a crucial role in training the models to achieve state-of-the-art performance on twelve tile-level tasks compared to the best-performing competitor models. While models that scale just in terms of parameter count tend to underperform when faced with diverse data and domain-specific training, it was found that domain-tailoring, data scale, and model scale contributed to better overall performance.
The team meticulously evaluated the performance of these foundation models on twelve tasks in the study, capturing the scope of computational pathology’s application areas. Preliminary results indicate that Virchow2 and Virchow2G excel at detecting minute details in the architecture and shapes of cells. Cell division detection and gene activity prediction are two areas where they excel. Complex characteristics, such as the direction and form of the cell nucleus, can probably be better measured for these jobs. This thorough evaluation should reassure the scientific and medical community about the reliability of these models in computational pathology and inspire optimism for the future of cancer diagnosis and treatment.Â
Check out the Paper and Blog. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter..
Don’t Forget to join our 48k+ ML SubReddit
Find Upcoming AI Webinars here
The post Microsoft and Paige Researchers Developed Virchow2 and Virchow2G: Second-Generation Foundation Models for Computational Pathology appeared first on MarkTechPost.
Source: Read MoreÂ