According to the National Cancer Institute, a cancer biomarker is a “biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or of a condition or disease such as cancer.†Biomarkers typically differentiate an affected patient from a person without the disease. Well-known cancer biomarkers include EGFR for lung cancer, HER2 for breast cancer, PSA for prostrate cancer, and so on. The BEST (Biomarkers, EndpointS, and other Tools) resource categorizes biomarkers into several types such as diagnostic, prognostic, and predictive biomarkers that can be measured with various techniques including molecular, imaging, and physiological measurements.
A study published in Nature Reviews Drug Discovery mentions that the overall success rate for oncology drugs from Phase I to approval is only around 5%. Biomarkers play a crucial role in enhancing the success of clinical development by improving patient stratification for trials, expediting drug development, reducing costs and risks, and enabling personalized medicine. For example, a study of 1,079 oncology drugs found that the success rates for drugs developed with a biomarker was 24% versus 6% for compounds developed without biomarkers.
Research scientists and real-world evidence (RWE) experts face numerous challenges to analyze biomarkers and validate hypotheses for biomarker discovery with their existing set of tools. Most notably, this includes manual and time-consuming steps for search, summarization, and insight generation across various biomedical literature (for example, PubMed), public scientific databases (for example, Protein Data Bank), commercial data banks and internal enterprise proprietary data. They want to quickly use, modify, or develop tools necessary for biomarker identification and correlation across modalities, indications, drug exposures and treatments, and associated endpoint outcomes such as survival. Each experiment might employ various combinations of data, tools, and visualization. Evidence in scientific literature should be simple to identify and cite with relevant context.
Amazon Bedrock Agents enables generative AI applications to automate multistep tasks by seamlessly connecting with company systems, APIs, and data sources. Bedrock multi-agent collaboration enables developers to build, deploy, and manage multiple specialized agents working together seamlessly to address increasingly complex business workflows. In this post, we show you how agentic workflows with Amazon Bedrock Agents can help accelerate this journey for research scientists with a natural language interface. We define an example analysis pipeline, specifically for lung cancer survival with clinical, genomics, and imaging modalities of biomarkers. We showcase a variety of specialized agents including a biomarker database analyst, statistician, clinical evidence researcher, and medical imaging expert in collaboration with a supervisor agent. We demonstrate advanced capabilities of agents for self-review and planning that help build trust with end users by breaking down complex tasks into a series of steps and showing the chain of thought to generate the final answer. The code for this solution is available in GitHub.
Multi-modal biomarker analysis workflow
Some example scientific requirements from research scientists analyzing multi-modal patient biomarkers include:
- What are the top five biomarkers associated with overall survival? Show me a Kaplan Meier plot for high and low risk patients.
- According to literature evidence, what properties of the tumor are associated with metagene X activity and EGFR pathway?
- Can you compute the imaging biomarkers for the patient cohort with low gene X expression? Show me the tumor segmentation and the sphericity and elongation values.
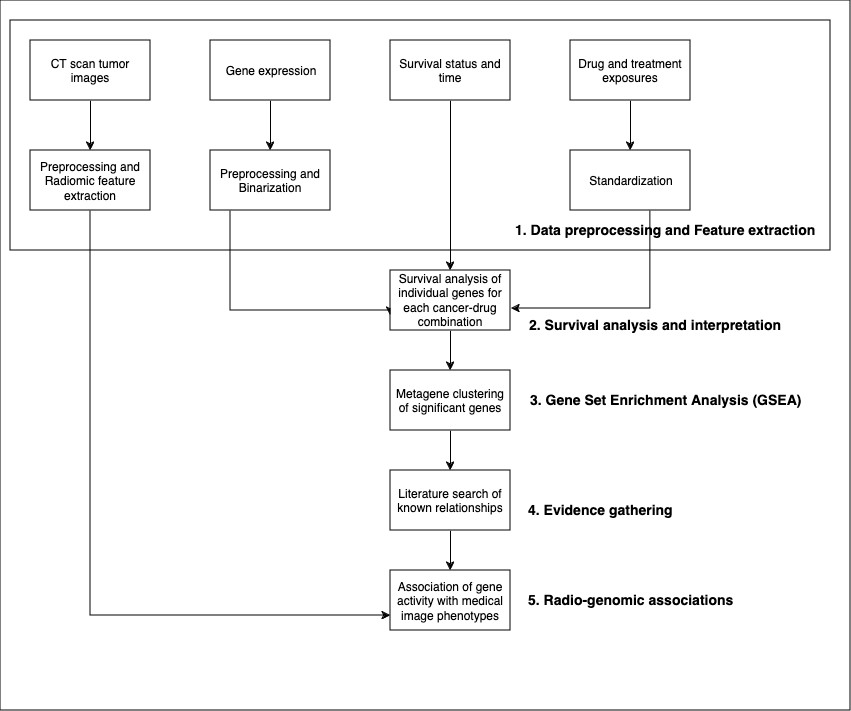
To answer the preceding questions, research scientists typically run a survival analysis pipeline (as shown in the following illustration) with multimodal data; including clinical, genomic, and computed tomography (CT) imaging data.
They might need to:
- Preprocess programmatically a diverse set of input data, structured and unstructured, and extract biomarkers (radiomic/genomic/clinical and others).
- Conduct statistical survival analyses such as the Cox proportional hazards model, and generate visuals such as Kaplan-Meier curves for interpretation.
- Conduct gene set enrichment analysis (GSEA) to identify significant genes.
- Research relevant literature to validate initial findings.
- Associate findings to radiogenomic biomarkers.

Solution overview
We propose a large-language-model (LLM) agents-based framework to augment and accelerate the above analysis pipeline. Design patterns for LLM agents, as described in Agentic Design Patterns Part 1 by Andrew Ng, include the capabilities for reflection, tool use, planning and multi-agent collaboration. An agent helps users complete actions based on both proprietary and public data and user input. Agents orchestrate interactions between foundation models (FMs), data sources, software applications, and user conversations. In addition, agents automatically call APIs to take actions and search knowledge bases to supplement information for these actions.

As shown in the preceding figure, we define our solution to include planning and reasoning with multiple sub-agents including:
- Biomarker database analyst: Convert natural language questions to SQL statements and execute on an Amazon Redshift database of biomarkers.
- Statistician: Use a custom container with lifelines library to build survival regression models and visualization such as Kaplan Meier charts for survival analysis.
- Clinical evidence researcher: Use PubMed APIs to search biomedical literature for external evidence. Use Amazon Bedrock Knowledge Bases for Retrieval Augmented Generation (RAG) to deliver responses from internal literature evidence.
- Clinical trial analyst: Use Clinicaltrials.gov APIs to search past clinical trial studies.
- Medical imaging expert: Use Amazon SageMaker jobs to augment agents with the capability to trigger asynchronous jobs with an ephemeral cluster to process CT scan images.
Dataset description
The non-small cell lung cancer (NSCLC) radiogenomic dataset comprises medical imaging, clinical, and genomic data collected from a cohort of early-stage NSCLC patients referred for surgical treatment. Each data modality presents a different view of a patient. It consists of clinical data reflective of electronic health records (EHR) such as age, gender, weight, ethnicity, smoking status, tumor node metastasis (TNM) stage, histopathological grade, and survival outcome. The genomic data contains gene mutation and RNA sequencing data from samples of surgically excised tumor tissue. It includes CT, positron emission tomography (PET)/CT images, semantic annotations of the tumors as observed on the medical images using a controlled vocabulary, segmentation maps of tumors in the CT scans, and quantitative values obtained from the PET/CT scans.
We reuse the data pipelines described in this blog post.
Clinical data
The data is stored in CSV format as shown in the following table. Each row corresponds to the medical records of a patient.
| Case ID | Survival status | Age at histological diagnosis | Weight (lbs) | Smoking status | Pack years | Quit smoking year | Chemotherapy | Adjuvant treatment | EGFR mutation status |
| R01-005 | Dead | 84 | 145 | Former | 20 | 1951 | No | No | Wildtype |
| R01-006 | Alive | 62 | Not collected | Former | Not collected | nan | No | No | Wildtype |
Genomics data
The following table shows the tabular representation of the gene expression data. Each row corresponds to a patient, and the columns represent a subset of genes selected for demonstration. The value denotes the expression level of a gene for a patient. A higher value means the corresponding gene is highly expressed in that specific tumor sample.
| Case_ID | LRIG1 | HPGD | GDF15 | CDH2 | POSTN |
| R01-024 | 26.7037 | 3.12635 | 13.0269 | 0 | 36.4332 |
| R01-153 | 15.2133 | 5.0693 | 0.90866 | 0 | 32.8595 |
Medical imaging data
The following image is an example overlay of a tumor segmentation onto a lung CT scan (case R01-093 in the dataset).

Deployment and getting started
Follow the deployment instructions described in the GitHub repo.
Full deployment takes approximately 10–15 minutes. After deployment, you can access the sample UI to test the agent with sample questions available in the UI or the chain of thought reasoning example.
The stack can also be launched in the us-east-1 or us-west-2 AWS Regions by choosing launch stack in the following:
| Region | codepipeline.yaml |
| us-east-1 | |
| us-west-2 |
Amazon Bedrock Agents deep dive
The following diagram describes the key components of the agent that interacts with the users through a web application.

Large language models
LLMs, such as Anthropic’s Claude or Amazon Titan models, possess the ability to understand and generate human-like text. They enable agents to comprehend user queries, generate appropriate responses, and perform complex reasoning tasks. In the deployment, we use Anthropic’s Claude 3 Sonnet model.
Prompt templates
Prompt templates are pre-designed structures that guide the LLM’s responses and behaviors. These templates help shape the agent’s personality, tone, and specific capabilities to understand scientific terminology. By carefully crafting prompt templates, you can help make sure that agents maintain consistency in their interactions and adhere to specific guidelines or brand voice. Amazon Bedrock Agents provides default prompt templates for pre-processing users’ queries, orchestration, a knowledge base, and a post-processing template.
Instructions
In addition to the prompt templates, instructions describe what the agent is designed to do and how it can interact with users. You can use instructions to define the role of a specific agent and how it can use the available set of actions under different conditions. Instructions are augmented with the prompt templates as context for each invocation of the agent. You can find how we define our agent instructions in agent_build.yaml.
User input
User input is the starting point for an interaction with an agent. The agent processes this input, understanding the user’s intent and context, and then formulates an appropriate chain of thought. The agent will determine whether it has the required information to answer the user’s question or need to request more information from the user. If more information is required from the user, the agent will formulate the question to request additional information. Amazon Bedrock Agents are designed to handle a wide range of user inputs, from simple queries to complex, multi-turn conversations.
Amazon Bedrock Knowledge Bases
The Amazon Bedrock knowledge base is a repository of information that has been vectorized from the source data and that the agent can access to supplement its responses. By integrating an Amazon Bedrock knowledge base, agents can provide more accurate and contextually appropriate answers, especially for domain-specific queries that might not be covered by the LLM’s general knowledge. In this solution, we include literature on non-small cell lung cancer that can represent internal evidence belonging to a customer.
Action groups
Action groups are collections of specific functions or API calls that Amazon Bedrock Agents can perform. By defining action groups, you can extend the agent’s capabilities beyond mere conversation, enabling it to perform practical, real-world tasks. The following tools are made available to the agent through action groups in the solution. The source code can be found in the ActionGroups folder in the repository.
- Text2SQL and Redshift database invocation: The Text2SQL action group allows the agent to get the relevant schema of the Redshift database, generate a SQL query for the particular sub-question, review and refine the SQL query with an additional LLM invocation, and finally execute the SQL query to retrieve the relevant results from the Redshift database. The action group contains OpenAPI schema for these actions. If the query execution returns a result greater than the acceptable lambda return payload size, the action group writes the data to an intermediate Amazon Simple Storage Service (Amazon S3) location instead.
- Scientific analysis with a custom container:Â The scientific analysis action group allows the agent to use a custom container to perform scientific analysis with specific libraries and APIs. In this solution, these include tasks such as fitting survival regression models and Kaplan Meier plot generation for survival analysis. The custom container allows a user to verify that the results are repeatable without deviations in library versions or algorithmic logic. This action group defines functions with specific parameters for each of the required tasks. The Kaplan Meier plot is output to Amazon S3.
- Biomedical literature evidence with PubMed: The PubMed action group allows the agent to interact with the PubMed Entrez Programming Utilities (E-utilities) API to fetch biomedical literature. The action group contains OpenAPI schema that accepts user queries to search across PubMed for articles. The Lambda function provides a convenient way to search for and retrieve scientific articles from the PubMed database. It allows users to perform searches using specific queries, retrieve article metadata, and handle the complexities of API interactions. Overall, the agent uses this action group and serves as a bridge between a researcher’s query and the PubMed database, simplifying the process of accessing and processing biomedical research information.
- Medical imaging with SageMaker jobs: The medical imaging action group allows the agent to process CT scan images of specific patient groups by triggering a SageMaker processing job. We re-use the medical imaging component from this previous blog.
The action group creates patient-level 3-dimensional radiomic features that explain the size, shape, and visual attributes of the tumors observed in the CT scans and stores them in Amazon S3. For each patient study, the following steps are performed, as shown in the figure that follows:
- Read the 2D DICOM slice files for both the CT scan and tumor segmentation, combine them to 3D volumes, and save the volumes in NIfTI format.
- Align CT volume and tumor segmentation so we can focus the computation inside the tumor.
- Compute radiomic features describing the tumor region using the pyradiomics library. It extracts 120 radiomic features of eight classes such as statistical representations of the distribution and co-occurrence of the intensity within the tumorous region of interest, and shape-based measurements describing the tumor morphologically.

Chain of thought reasoning and responses
Let’s look at a few examples of chain of thought reasoning and execution with the supervisor agent. The supervisor agent interprets the user question, generates a sequence of steps, and executes them with the relevant sub-agents.
To respond to the following questions, the agent sets up the following orchestration workflows with the available sub-agents.
Question: What is the best gene expression biomarker (lowest p value) with overall survival for patients that have undergone chemotherapy, show me a bar chart with the top five biomarkers.
- I will generate a plan
- Query the biomarker database for patients’ data
- Run survival analysis to find p-values
- Get the top 5 biomarkers with lowest p-values and create a visualization
- I will ask the Biomarker database analyst
- Get DB schema and generate SQL
- Review and refine the SQL with self-evaluation
- Execute the SQL query and store the results in Amazon S3
- I will ask the Statistician
- Retrieve the generated dataset from the database analyst
- Fit a survival regression model with the custom container API
- Retrieve the model fit summary results and sort the top five biomarkers based on the lowest p-value
- Generate a bar chart with the top five biomarkers
Supervisor response: Gene biomarker ‘gdf15’ has the lowest p value of 0.02, therefore gdf15 is the best gene biomarker. The bar chart shows the top 5 biomarkers and their corresponding p values.
Question: According to literature evidence, what properties of the tumor are associated with metagene X activity and EGFR pathway?
- I will generate a plan
- Route the request to the Clinical evidence researcher
- I will ask the Clinical evidence researcher
- Query internal knowledge base for internal evidence
- Query PubMed for external literature
- Summarize the results with citations
Supervisor response: Based on internal and external evidence, imaging properties, molecular characteristics, drug resistance and therapeutic approaches, signalling pathways and mechanisms, emerging therapeutic strategies , and cellular properties are relevant for the EGFR pathway.
Question: Can you compute the imaging biomarkers for patient cohort with low gene X expression? Show me the tumor segmentation and the sphericity and elongation values.
- I will generate a plan
- Query the biomarker database to find the patient cohort with lowest gdf15 values
- Trigger medical imaging analysis jobs to process CT scan images of the cohort
- Summarize the findings of the imaging jobs.
- I will ask the Biomarker database analyst
- Get DB schema and generate SQL
- Review and refine the SQL with self-evaluation
- Execute the SQL query and retrieve patient ids
- I will ask the medical imaging expert
- Retrieve the patient cohort from the database analyst
- Compute imaging biomarkers for each patient in the cohort with a SageMaker medical imaging job
- On job completion notification, retrieve and analyse the results
- Extract the required biomarkers and visualize the tumour segmentation output images
Supervisor response: The compute imaging biomarker job to process the medical images of the patients was triggered and is now complete. The following patients were selected based on their gene X expression values. The elongation and sphericity values for the patient tumours are displayed along with the corresponding tumour segmentation images.
Conclusion
Research scientists face significant challenges in identifying and validating biomarkers specific to cancer subtypes and relevant to interventions and patient outcomes. Existing tools often require intensive manual steps to search, summarize, and generate insights across diverse data sources. This post has demonstrated how Amazon Bedrock Agents can offer a flexible framework with multi-agent collaboration and relevant tools to help accelerate this critical discovery process.
By providing an example analysis pipeline for lung cancer survival, we showcased how these agentic workflows use a natural language interface, database retrieval, statistical modeling, literature search, and medical image processing to transform complex research queries into actionable insights. The agent used advanced and intelligent capabilities such as self-review and planning, breaking down tasks into step-by-step analyses and transparently displaying the chain of thought behind the final answers. While the potential impact of this technology on pharmaceutical research and clinical trial outcomes remains to be fully realized, solutions like this can help automate data analysis and hypothesis validation tasks.
The code for this solution is available on GitHub, and we encourage you to explore and build upon this template. For examples to get started with Amazon Bedrock Agents, check out the Amazon Bedrock Agents GitHub repository.
About the authors
 Hasan Poonawala is a Senior AI/ML Solutions Architect at AWS, working with Healthcare and Life Sciences customers. Hasan helps design, deploy and scale Generative AI and Machine learning applications on AWS. He has over 15 years of combined work experience in machine learning, software development and data science on the cloud. In his spare time, Hasan loves to explore nature and spend time with friends and family.
Hasan Poonawala is a Senior AI/ML Solutions Architect at AWS, working with Healthcare and Life Sciences customers. Hasan helps design, deploy and scale Generative AI and Machine learning applications on AWS. He has over 15 years of combined work experience in machine learning, software development and data science on the cloud. In his spare time, Hasan loves to explore nature and spend time with friends and family.
 Michael Hsieh is a Principal AI/ML Specialist Solutions Architect. He works with HCLS customers to advance their ML journey with AWS technologies and his expertise in medical imaging. As a Seattle transplant, he loves exploring the great mother nature the city has to offer, such as the hiking trails, scenery kayaking in the SLU, and the sunset at Shilshole Bay.
Michael Hsieh is a Principal AI/ML Specialist Solutions Architect. He works with HCLS customers to advance their ML journey with AWS technologies and his expertise in medical imaging. As a Seattle transplant, he loves exploring the great mother nature the city has to offer, such as the hiking trails, scenery kayaking in the SLU, and the sunset at Shilshole Bay.
 Nihir Chadderwala is a Senior AI/ML Solutions Architect on the Global Healthcare and Life Sciences team. His background is building big data and AI-powered solutions to customer problems in a variety of domains such as software, media, automotive, and healthcare. In his spare time, he enjoys playing tennis, and watching and reading about Cosmos.
Nihir Chadderwala is a Senior AI/ML Solutions Architect on the Global Healthcare and Life Sciences team. His background is building big data and AI-powered solutions to customer problems in a variety of domains such as software, media, automotive, and healthcare. In his spare time, he enjoys playing tennis, and watching and reading about Cosmos.
 Zeek Granston is an Associate AI/ML Solutions Architect focused on building effective artificial intelligence and machine learning solutions. He stays current with industry trends to deliver practical results for clients. Outside of work, Zeek enjoys building AI applications, and playing basketball.
Zeek Granston is an Associate AI/ML Solutions Architect focused on building effective artificial intelligence and machine learning solutions. He stays current with industry trends to deliver practical results for clients. Outside of work, Zeek enjoys building AI applications, and playing basketball.
Source: Read MoreÂ