Recent advancements in SSL have led to the development of foundation models (FMs) that analyze extensive biomedical data, enhancing health outcomes. Continuous Glucose Monitoring (CGM) offers rich, temporal glucose data but must be utilized for broader health predictions. SSL enables FMs to analyze unlabelled data efficiently, improving detection rates in various medical fields, from retinal images to sleep disorders and pathology. With diabetes affecting over 500 million people globally and increasing healthcare costs, CGM has proven superior to traditional glucose monitoring by improving glycemic control and quality of life. The FDA’s approval of an over-the-counter CGM device highlights its growing accessibility and potential benefits for diabetic and non-diabetic individuals.
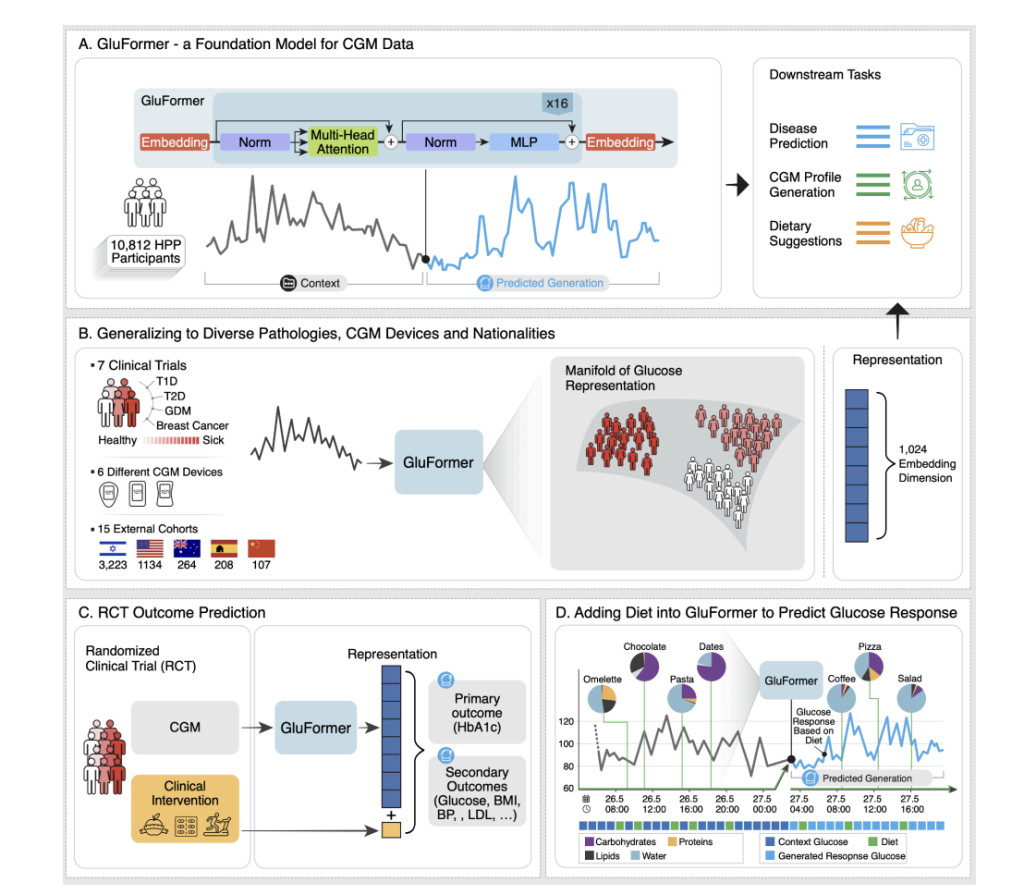
Researchers from institutions including Weizmann Institute of Science and NVIDIA introduce GluFormer, a generative foundation model using a transformer architecture, trained on over 10 million CGM measurements from 10,812 non-diabetic individuals. GluFormer, trained with a self-supervised approach, generalizes well across 15 external datasets and various populations, showing superior performance in predicting clinical parameters such as HbA1c, liver metrics, and future health outcomes, including up to four years ahead. The model also integrates dietary data to simulate dietary interventions and personalize food responses. GluFormer represents a significant advancement in managing chronic diseases and improving precision health strategies.
GluFormer is a transformer-based model designed to analyze CGM data from 10,812 participants; each tracked for two weeks. The model processes glucose readings, which are recorded every 15 minutes and initially cleansed of calibration noise. Data was tokenized into 460 glucose value intervals and segmented into 1200-measurement samples, with a special token used for shorter samples. The model architecture includes 16 transformer layers, 16 attention heads, and an embedding dimension of 1024, handling sequences of up to 25,000 tokens. Pretraining involved predicting subsequent tokens using causal masking and cross-entropy loss. Optimization was performed with AdamW and a learning rate scheduler, and the final evaluation was based on performance metrics from a validation set.
GluFormer outputs were pooled for clinical application using methods such as Max Pooling, which proved most effective for predicting clinical outcomes like HbA1c. Clinical metrics derived from CGM data using the R package iglu were used with ridge regression models to predict various outcomes. The model’s generalizability was assessed on external datasets, and its ability to predict outcomes of randomized clinical trials was evaluated. Comparisons were made with other models, including a plain transformer and CNNs. At the same time, methods for incorporating temporal information and dietary data were also explored, demonstrating that learned temporal embeddings and diet tokens improved predictive performance.
The GluFormer model demonstrated robust performance in generating CGM data and predicting clinical outcomes. Trained on a large dataset, GluFormer excels in producing accurate CGM signals and embedding them for various applications. It showed strong performance in predicting clinical parameters like HbA1c, visceral adipose tissue, and fasting glucose. The model effectively generalizes across cohorts and integrates dietary data, enhancing glucose response predictions. Improvements, including temporal encoding and multimodal integration, refine its predictive accuracy. These results underscore GluFormer’s utility in personalized healthcare and its adaptability to diverse datasets and timeframes.
In conclusion, GluFormer, trained on CGM data from over 10,000 non-diabetic individuals, excels in generating accurate glucose signals and predicting a range of clinical outcomes. It outperforms traditional metrics in forecasting parameters like HbA1c and liver function and shows broad applicability across different populations and health conditions. The model’s latent space effectively captures nuanced aspects of glucose metabolism, enhancing its ability to predict diabetes progression and clinical trial outcomes. Integration of dietary data further refines its predictions. Despite its potential, challenges remain, including dataset limitations, dietary data accuracy, and model interpretability. GluFormer represents a significant advancement in personalized metabolic health management and research.
Check out the Paper and Project. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter..
Don’t Forget to join our 50k+ ML SubReddit
The post GluFormer: Advancing Personalized Metabolic Health through Generative AI Modeling and Self-Supervised Learning appeared first on MarkTechPost.
Source: Read MoreÂ