The integration of language models into biological research represents a significant challenge due to the inherent differences between natural language and biological sequences. Biological data, such as DNA, RNA, and protein sequences, are fundamentally different from natural language text, yet they share sequential characteristics that make them amenable to similar processing techniques. The primary challenge lies in effectively adapting language models, originally developed for natural language processing (NLP), to handle the complexities of biological sequences. Addressing this challenge is crucial for enabling more accurate predictions in fields such as protein structure prediction, gene expression analysis, and the identification of molecular interactions. Successfully overcoming these hurdles has the potential to revolutionize various domains within biology, particularly in areas requiring the analysis of large and complex datasets.
Current methods for analyzing biological sequences rely heavily on traditional sequence alignment techniques and machine learning approaches. Sequence alignment tools like BLAST and Clustal are commonly used but often struggle with the computational complexity and scalability required for large datasets. These methods are further limited by their inability to capture the deeper structural and functional relationships within sequences. Machine learning techniques, including random forests and support vector machines, offer some improvements but are constrained by the need for manually engineered features and their lack of generalizability across diverse biological contexts. These limitations significantly reduce the effectiveness and applicability of these methods, particularly in real-time biological research where efficiency and accuracy are paramount.
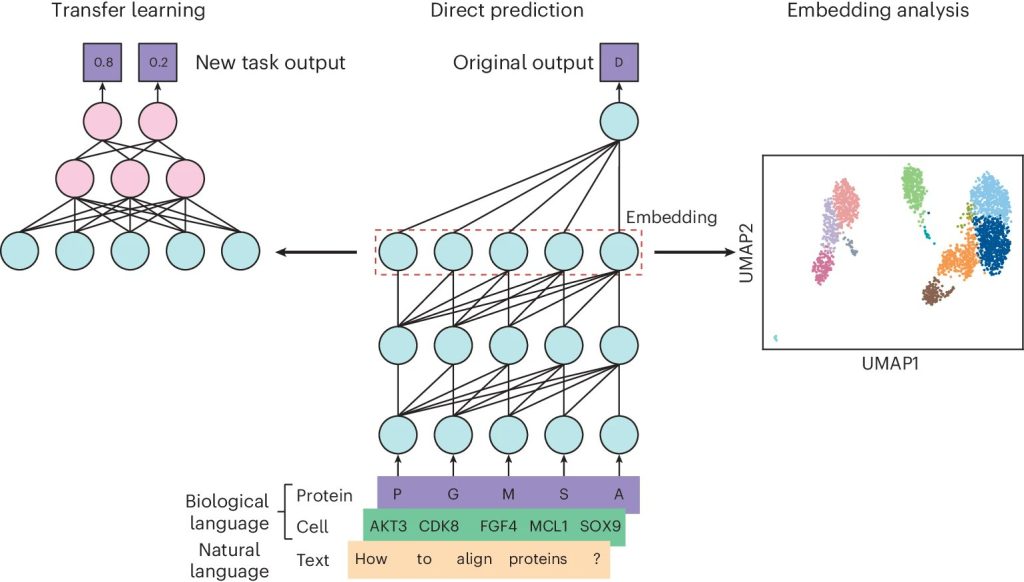
To address these limitations, Stanford researchers propose using language models, particularly those based on the transformer architecture, in biological research. This innovative approach leverages the ability of language models to process large-scale, heterogeneous datasets and to uncover complex patterns within sequential data. Pre-trained language models, such as ESM-2 for protein sequences and Geneformer for single-cell data, can be fine-tuned for specific biological tasks, offering a flexible and scalable solution that addresses the shortcomings of traditional methods. By harnessing the power of these models, the approach provides a significant advancement in the analysis of biological sequences, enabling more accurate and efficient predictions in critical areas of research.
The proposed method relies on the transformer architecture, which is particularly effective for processing sequential data. The researchers have utilized various pre-trained models, including ESM-2, a protein language model trained on over 250 million protein sequences, and Geneformer, a single-cell language model trained on 30 million single-cell transcriptomes. These models employ masked language modeling, where parts of the sequence are hidden, and the model is trained to predict the missing elements. This training enables the model to learn the underlying patterns and relationships within the sequences, making it possible to predict outcomes such as protein stability, gene expression levels, and variant effects. The models can be further fine-tuned for specific tasks, such as integrating multi-modal data that includes gene expression, chromatin accessibility, and protein abundance.
The proposed language models demonstrated substantial improvements across various biological tasks. For protein sequence analysis, the model achieved higher accuracy in predicting protein stability and evolutionary constraints, significantly outperforming existing methods. In single-cell data analysis, the model effectively predicted cell types and gene expression patterns with enhanced precision, offering superior performance in identifying subtle biological variations. These results underscore the models’ potential to transform biological research by providing accurate, scalable, and efficient tools for analyzing complex biological data, thereby advancing the capabilities of computational biology.
In conclusion, this proposed method offers a significant contribution to AI-driven biological research by effectively adapting language models for the analysis of biological sequences. The approach addresses a critical challenge in the field by leveraging the strengths of transformer-based models to overcome the limitations of traditional methods. The use of models like ESM-2 and Geneformer provides a scalable and accurate solution for a wide range of biological tasks, with the potential to revolutionize fields such as genomics, proteomics, and personalized medicine by enhancing the efficiency and accuracy of biological data analysis.
Check out the Paper and Colab Tutorial. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter..
Don’t Forget to join our 48k+ ML SubReddit
Find Upcoming AI Webinars here
The post LLM for Biology: This Paper Discusses How Language Models can be Applied to Biological Research appeared first on MarkTechPost.
Source: Read MoreÂ