Recent advances in immune sequencing and experimental methods generate extensive T cell receptor (TCR) repertoire data, enabling models to predict TCR binding specificity. T cells play a role in the adaptive immune system, orchestrating targeted immune responses through TCRs that recognize non-self antigens from pathogens or diseased cells. TCR diversity, essential for recognizing diverse antigens, is generated through random DNA rearrangement involving V, D, and J gene segments. While theoretical TCR diversity is extremely high, the actual diversity in an individual is much smaller. TCRs interact with peptides on the major histocompatibility complex (pMHC), with some TCRs recognizing numerous pMHC complexes.
Researchers from IBM Research Europe, the Institute of Computational Life Sciences at Zürich University of Applied Sciences, and Yale School of Medicine review the evolution of computational models for predicting TCR binding specificity. Emphasizing machine learning, they cover early unsupervised clustering approaches, supervised models, and the transformative impact of Protein Language Models (PLMs) in bioinformatics, particularly in TCR specificity analysis. The review addresses dataset biases, generalization issues, and model validation shortcomings. It highlights the importance of improving model interpretability and extracting biological insights from large, complex models to enhance TCR-pMHC binding predictions and revolutionize immunotherapy development.
TCR specificity data comes from databases like VDJdb and McPas-TCR, but these datasets have significant limitations. Bulk sequencing is high-throughput and cost-effective but can’t detect paired α and β chains, while single-cell technologies that can are expensive and underrepresented. Most datasets focus on a limited number of epitopes, predominantly of viral origin and associated with common HLA alleles, showing significant bias. Additionally, the lack of negative data complicates supervised machine learning model development. Generating artificial negative pairs introduces biases, and high-performance models can memorize sequences, leading to over-optimistic results. Ensuring generated negative pairs accurately reflect true non-binding distributions remains a challenge.
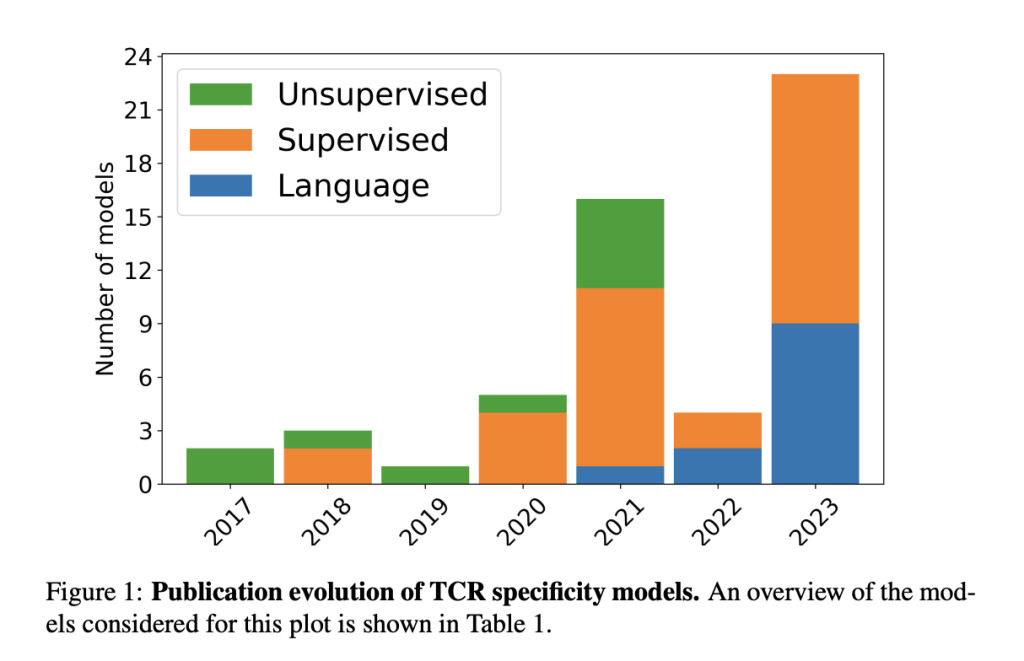
Since 2017, the modeling of TCR specificity has evolved significantly, beginning with unsupervised clustering methods. Initial models like TCRdist and GLIPH grouped TCRs based on sequence similarities and biochemical properties. These methods demonstrated that TCR sequences contain valuable specificity information, but they struggled with complex nonlinear interactions. This prompted the development of supervised models that utilized machine learning techniques to handle the increasing complexity of data better. Early supervised models, including TCRGP and TCRex, employed classifiers such as Gaussian Processes and random forests to predict TCR specificity. Meanwhile, neural network-based approaches like NetTCR and DeepTCR leveraged advanced architectures to enhance predictive accuracy.
The introduction of PLMs marked the latest advancement in TCR specificity prediction. Based on Transformer architectures, these models were trained on extensive protein sequence datasets, achieving remarkable performance in various protein-related tasks. TCR-BERT and STAPLER, for example, utilized BERT-based models fine-tuned for TCR and antigen classification, demonstrating the effectiveness of PLMs in capturing complex sequence interactions. Despite their success, challenges remain in addressing lexical ambiguity and enhancing model interpretability. Future improvements in embedding optimization and adaptation of interpretability methods specific to protein sequences are crucial for further advancements in TCR specificity prediction.
Accurate TCR specificity prediction is vital for improving immunotherapies and understanding autoimmune diseases. Limited and biased data, particularly epitope information, challenge current models, hindering generalization to new epitopes. Advances in machine learning, including CNNs, RNNs, transfer learning, and PLMs, have significantly enhanced TCR prediction models, but challenges remain, especially in predicting specificity for novel epitopes. Benchmarks like IMMREP22 and IMMREP23 highlight difficulties in fair model comparison and generalizability. Adapting TCR models for BCR prediction, which involves non-linear epitopes and complex antigen interactions, presents further computational challenges.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter..
Don’t Forget to join our 47k+ ML SubReddit
Find Upcoming AI Webinars here
The post Advances and Challenges in Predicting TCR Specificity: From Clustering to Protein Language Models appeared first on MarkTechPost.
Source: Read MoreÂ