Recently, there has been remarkable performance on clinical question-answer (QA) tasks by large language models (LLMs) like Med-PaLM 2 and GPT-4. For example, Med-PaLM 2 produced answers to consumer health questions that were competitive with human doctors, and a GPT-4-based system achieved 90.2% on the MedQA task. But these models have a lot of problems. They are costly to train and run and ecologically unsustainable because their parameter counts can reach into the billions, necessitating dedicated computing clusters. Researchers can only access these large models through a paid API. Researchers and practitioners are thus unable to analyze these models, and only individuals having access to the model’s weights and architecture can research improvements.
A new and promising approach, known as on-device AI or edge AI, utilizes local devices like phones or tablets to run language models. This technology holds immense potential in biomedicine, offering solutions such as disseminating medical information after catastrophic events or in areas with limited or no internet service. Despite the challenges posed by their size and closed nature, models like GPT-4 and Med-PaLM 2 can be adapted for on-device AI, opening up new avenues for research and application in the field.
In a biomedical context, two types of models are applicable. Only biomedical text from PubMed was used to train smaller domain-specific models (<3B parameters) like BioGPT-large and BioMedLM. Larger 7B parameter models like LLaMA 2 and Mistral 7B are more powerful than their smaller counterparts. However, they were trained on broad English text and did not have a biological focus. How well these models work and which is best suited for clinical QA applications are still in the air.
To ensure comprehensive and reliable findings, a team of researchers from Stanford University, University College London, and the University of Cambridge conducted a rigorous evaluation of all four models in the clinical QA domain. They used two popular tasks, MedQA (questions similar to those on the USMLE) and MultiMedQA Long Form Answering (open response to consumer health queries), which assess the ability to understand and reason about medical scenarios and write informative paragraphs responding to health questions.
The MedQA four-option activity is similar to the USMLE in that it asks a question with four possible answers. This test commonly assesses a language model’s ability to use medical information and reason about clinical situations. Some questions may seek particular medical information (such as schizophrenia symptoms), while others may pose a clinical scenario and ask for the best diagnosis or next step (such as, “A 27-year-old male presents… “).Â
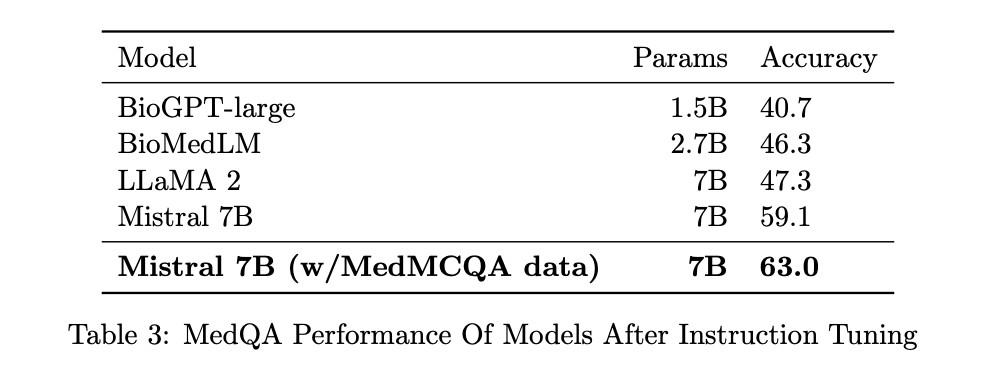
There are 1273 test cases, 10178 training instances, and 1272 development examples in the MedQA dataset. A prompt and an expected response were provided for each example. The four models were taught to use the same prompt and offer the same response, just the word “Answer: “accompanied by the letter representing the right choice. Comparison of Four-Way Models By updating all of their parameters, all four models were fine-tuned using the 10178 training instances. The researchers used the same format, training data, and training code for all the models to ensure they could compare them fairly. To get the models just right, they used the Hugging Face package.
By merging the MedQA training data with the bigger MedMCQA training set, which includes 182822 more examples, the top-performing model (Mistral 7B) was fine-tuned, allowing researchers to delve deeper into the capabilities of mid-size models. Research has demonstrated that using this data for training improves MedQA performance. At this stage, they trained the model to produce the right letter and the complete text of the response using a somewhat more complex request. A comparable hyperparameter sweep was used to find the optimal values. Remember that the primary goal of these trials was to optimize Mistral 7B’s performance rather than to provide an accurate evaluation of competing models.
To train the model for the MultiMedQA Long Form Question Answering job, the researchers fed it health-related queries that users often submit to search engines. Three datasets—LiveQA, MedicationQA, and HealthSearchQA—contribute to the four thousand questions. LiveQA also includes answers to frequently asked questions. Similarly to a response to a health-related frequently asked questions page, the system is anticipated to produce a detailed response of one or two paragraphs. The comprehensive set of questions covers infectious diseases, chronic illnesses, dietary deficiencies, reproductive health, developmental issues, drug usage, pharmaceutical interactions, preventative measures, and a host of other consumer health subjects.
These findings have practical implications for the field of biomedicine. Mistral 7 B emerged as the top performer on both tests, demonstrating its potential for clinical question-answering tasks. BioMedLM, while less bulky than the 7B versions, also showed respectable performance. For those with the computational resources, BioGPT-large can provide satisfactory results. However, the researchers noted that domain-specific models performed worse on both tasks than larger-scale models trained on generic English, which might have incorporated the PubMed corpus. The question of whether a larger biomedical specialty model would significantly outperform Mistral 7B remains open, highlighting the need for expert medical review of model outputs before their clinical application. Â
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our Telegram Channel, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 40k+ ML SubReddit
The post Researchers at Stanford Explore the Potential of Mid-Sized Language Models for Clinical QA (Question-Answering) Tasks appeared first on MarkTechPost.
Source: Read MoreÂ