Sleep medicine is a critical field that involves monitoring and evaluating physiological signals to diagnose sleep disorders and understand sleep patterns. Techniques such as polysomnography (PSG) record brain, cardiac, and respiratory activities during sleep, providing a detailed overview of a person’s sleep health. These signals are essential in categorizing sleep stages and identifying sleep disorders. PSG typically includes electroencephalograms (EEG), electrooculograms (EOG), electromyograms (EMG), electrocardiograms (ECG), and respiratory channels. Each modality offers a unique perspective: brain activity signals (BAS) measure brain function, ECG monitors heart rhythms, and respiratory sensors quantify breathing patterns, collectively providing a comprehensive assessment of sleep health.
Accurately analyzing sleep data is crucial due to the complexity of sleep disorders. Manual analysis, which involves visual inspection by trained technicians, is time-consuming, labor-intensive, and prone to errors. This traditional method faces significant challenges, especially with the increasing volume of sleep data. Therefore, there is a pressing need for automated techniques that can efficiently and accurately analyze sleep data across multiple physiological signals. The goal is to develop robust models that can handle the complexity of sleep data and provide reliable diagnoses.
Current methods for sleep data analysis primarily rely on supervised deep-learning models. These models have shown promise in automating sleep staging and the classification of sleep disorders like sleep-disordered breathing (SDB). However, most existing methods depend on labeled data from narrow tasks and do not leverage the full breadth of physiological signals available from PSG. For instance, DL models such as CNNs and RNNs have been proposed for sleep-scoring tasks but often need to catch up in generalizability and robustness. Additionally, while contrastive learning (CL) has been successful in other domains, its application in integrating BAS, ECG, and respiratory signals for sleep analysis remains underexplored.
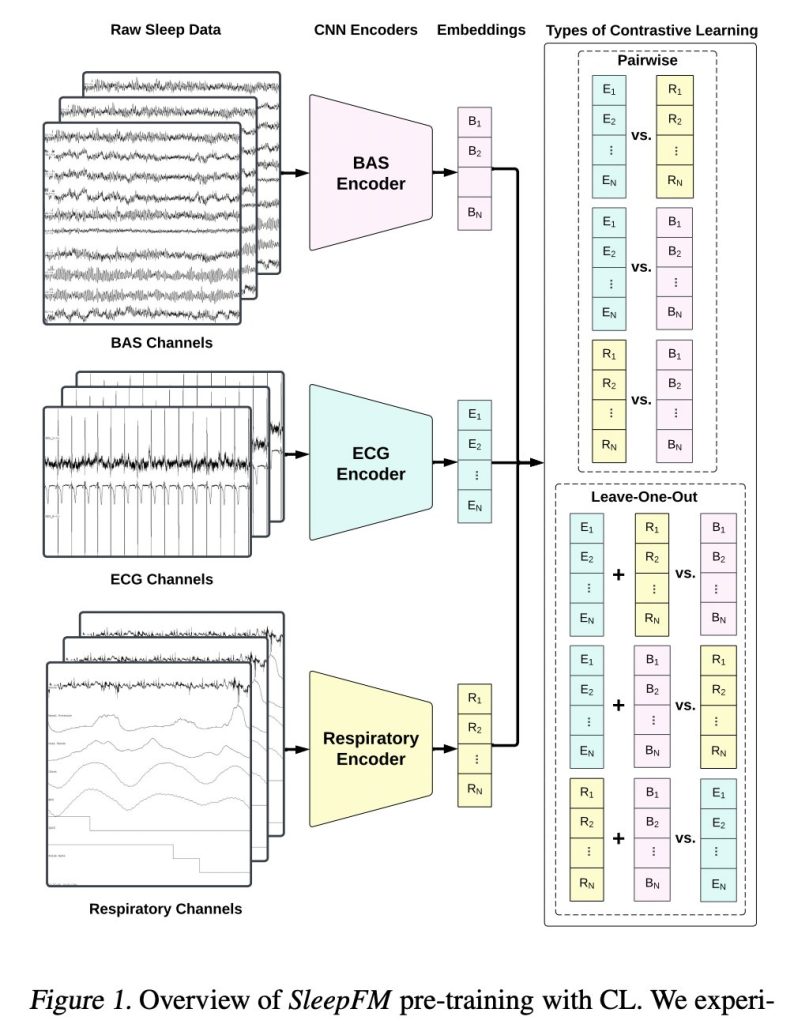
Researchers from Stanford University and the Technical University of Denmark introduced SleepFM, a groundbreaking multi-modal foundation model for sleep analysis. This model leverages a vast dataset of multi-modal sleep recordings from over 14,000 participants, totaling more than 100,000 hours of sleep data collected between 1999 and 2020 at the Stanford Sleep Clinic. SleepFM utilizes a contrastive learning approach to integrate brain activity, ECG, and respiratory signals. This integration enables the model to capture comprehensive physiological representations, significantly enhancing the accuracy of sleep analysis.
SleepFM employs three 1D convolutional neural networks (CNNs) to generate embeddings from each modality (BAS, ECG, and respiratory signals). The architecture of these models is based on a 1D CNN developed for classifying ECG measurements. Each CNN is tailored to handle the specific characteristics of its respective modality: 10 channels for BAS, 2 for ECG, and 7 for respiratory channels. A novel leave-one-out contrastive learning technique is introduced, significantly outperforming the standard pairwise contrastive learning in capturing the synergy between different physiological signals.
In sleep stage classification, SleepFM achieved a macro AUROC of 0.88 and a macro AUPRC of 0.72, compared to 0.72 and 0.48 by end-to-end CNNs. SleepFM outperformed CNNs with an AUROC of 0.85 and an AUPRC of 0.77 for sleep-disordered breathing detection, versus 0.69 and 0.61 by CNNs. Furthermore, SleepFM’s embeddings demonstrated a 48% top-1 average accuracy in retrieving corresponding recording clips of other modalities from 90,000 candidates. These results underscore the model’s ability to integrate diverse physiological signals and improve the accuracy and efficiency of sleep analysis.
The model’s success is mostly attributed to its ability to learn rich, multi-modal representations of physiological data, which are crucial for accurate sleep analysis. SleepFM also excelled in demographic attributes classification, showing high accuracy in predicting age and gender from 30-second clips of physiological data. The model achieved AUROCs of 0.982, 0.852, 0.784, and 0.915 for the age groups 0-18, 18-35, 35-50, and 50+, respectively. For gender classification, the AUROC was 0.850, significantly outperforming baseline models.
In conclusion, SleepFM represents significant progress in sleep medicine by providing an automated, accurate, and efficient method for analyzing multi-modal sleep data. SleepFM offers a holistic approach to understanding sleep patterns and diagnosing disorders by integrating brain activity, ECG, and respiratory signals. The model’s superior performance across various tasks, including sleep stage classification, sleep-disordered breathing detection, and demographic prediction, highlights its potential to transform clinical practices in sleep medicine. The success of SleepFM demonstrates the value of holistic multi-modal sleep modeling in capturing the richness of sleep recordings, ultimately contributing to better understanding and improving sleep health.
Check out the Paper and GitHub. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our Telegram Channel, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 43k+ ML SubReddit | Also, check out our AI Events Platform
The post Researchers at Stanford Propose SleepFM: A New Multi-Modal Foundation Model for Sleep Analysis appeared first on MarkTechPost.
Source: Read MoreÂ