

Autoregressive protein language models (pLMs) have become transformative tools for designing functional proteins with remarkable diversity, demonstrating success in creating enzyme families like lysozymes and carbonic anhydrases. These models generate protein sequences by sampling from learned probability distributions, uncovering intrinsic patterns within training datasets. Despite their ability to explore high-quality subspaces of the sequence landscape, pLMs struggle to target rare and valuable regions, limiting their effectiveness in tasks like engineering enzymatic activity or binding affinity. This challenge, compounded by the vast sequence space and expensive wet lab validation, makes protein optimization a complex problem. Traditional methods like directed evolution, which iteratively select desired traits, are limited to local exploration and lack tools for steering long-term evolutionary trajectories toward specific biological functions.
RL offers a promising framework to guide pLMs toward optimizing specific properties by aligning model outputs with feedback from an external oracle, such as predicted stability or binding affinities. Drawing inspiration from RL applications in robotics and gaming, recent efforts have applied RL techniques to protein design, demonstrating the potential to explore rare events and balance exploration-exploitation trade-offs efficiently. Examples include Proximal Policy Optimization (PPO) for DNA and protein design and Direct Preference Optimization (DPO) for thermostability prediction and binder design. While these studies showcase RL’s potential, there remains a need for experimentally validated, publicly available RL frameworks tailored to generative pLMs, which could advance the field of protein engineering.
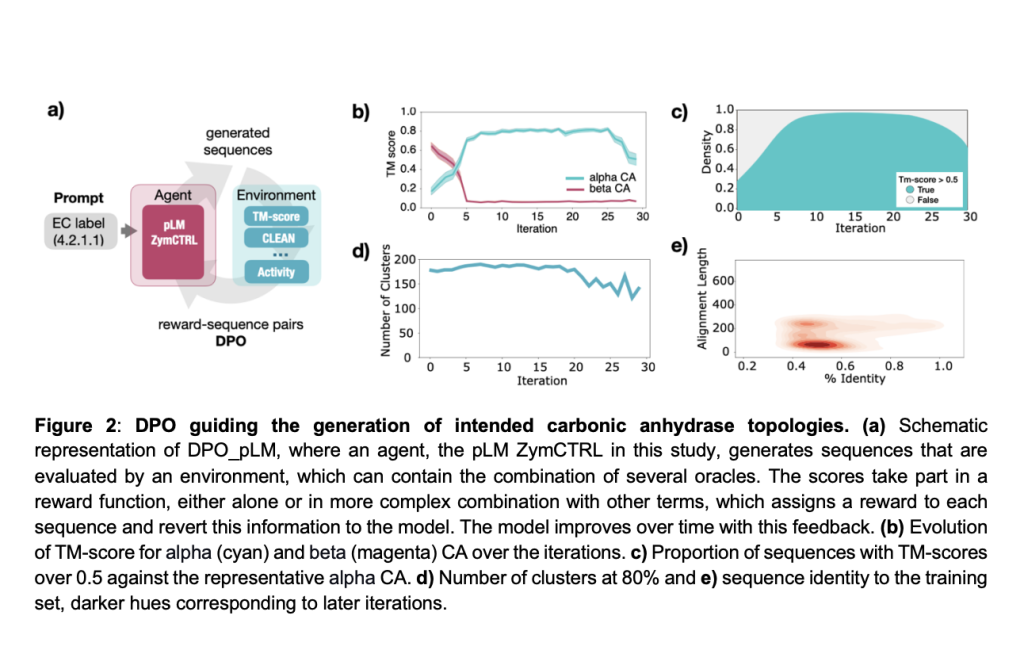
Researchers from Universitat Pompeu Fabra, the Centre for Genomic Regulation, and other leading institutions developed DPO_pLM, an RL framework for optimizing protein sequences with generative pLMs. By fine-tuning pLMs using rewards from external oracles, DPO_pLM optimizes diverse user-defined properties without additional data while preserving sequence diversity. It outperforms traditional fine-tuning methods by reducing computational demands, mitigating catastrophic forgetting, and leveraging negative data. Demonstrating its effectiveness, DPO_pLM successfully designed nanomolar-affinity EGFR binders within hours.
The study introduces DPO and self-fine-tuning (s-FT) for optimizing protein sequences. DPO minimizes loss functions, including ranked and weighted forms, with negative log-likelihood proving effective. s-FT refines ZymCTRL iteratively, generating, ranking, and fine-tuning top sequences across 30 iterations. Model training uses Hugging Face’s transformers API, employing batch sizes of 4, a learning rate of 8×10⁻⁶, and evaluation every 10 steps. Structural similarity is assessed using ESMFold and Foldseek, while functional annotations rely on ESM1b embeddings and cosine similarity with CLEAN clusters. EGFR binder design applies fine-tuning on BLAST-retrieved sequences, followed by AlphaFold folding and optimization to enhance binder performance.
pLMs generate sequences resembling their training data and often achieve high functionality despite significant sequence deviations. For instance, ZymCTRL, trained on enzyme data with EC labels, created carbonic anhydrases with wild-type activity but only 39% sequence identity. Similarly, generated α-amylases outperformed wild-type activity. However, pLMs primarily replicate training set distributions, lacking precise control for optimizing specific properties like activity or stability. By applying RL, particularly methods like DPO, pLMs can be fine-tuned iteratively using feedback from oracles, enabling the generation of sequences with targeted properties while preserving diversity and quality.
In conclusion, pLMs excel at sampling from distributions but struggle to optimize specific properties. DPO_pLM overcomes this limitation by utilizing Direct Preference Optimization DPO, which refines sequences through external oracles without additional training data. ZymCTRL evaluations showed rapid and robust performance, enriching enzyme classes and folds in multi-objective tasks. In an EGFR binder design experiment, DPO_pLM achieved a 50% success rate, generating three nanomolar binders after 12 iterations in just hours. Unlike fine-tuning, DPO maximizes preference rewards, improving global predictions efficiently. Future work will focus on integrating DPO_pLM into automated labs for protein design innovations.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. Don’t Forget to join our 60k+ ML SubReddit.
The post Optimizing Protein Design with Reinforcement Learning-Enhanced pLMs: Introducing DPO_pLM for Efficient and Targeted Sequence Generation appeared first on MarkTechPost.
Source: Read MoreÂ


 Trending: LG AI Research Releases EXAONE 3.5: Three Open-Source Bilingual Frontier AI-level Models Delivering Unmatched Instruction Following and Long Context Understanding for Global Leadership in Generative AI Excellence….
Trending: LG AI Research Releases EXAONE 3.5: Three Open-Source Bilingual Frontier AI-level Models Delivering Unmatched Instruction Following and Long Context Understanding for Global Leadership in Generative AI Excellence….