

Cell segmentation and classification are vital tasks in spatial omics data analysis, which provides unprecedented insights into cellular structures and tissue functions. Recent advancements in spatial omics technologies have enabled high-resolution analysis of intact tissues, supporting initiatives like the Human Tumor Atlas Network and the Human Biomolecular Atlas Program in mapping spatial organizations in healthy and diseased states. Traditional workflows treat segmentation and classification as separate steps, relying on CNN-based methods like Mesmer, Cellpose, and CELESTA. However, these approaches often need more computational efficiency, consistent performance across tissue types, and a lack of confidence assessment in segmentation, necessitating advanced computational solutions.
Although CNNs have improved biomedical image segmentation and classification, their limitations hinder semantic information integration within tissue images. Transformer-based models, such as DETR, DINO, and MaskDINO, outperform CNNs in object detection and segmentation tasks, showing promise for biomedical imaging. Yet, their application to cell and nuclear segmentation in multiplexed tissue images still needs to be explored. Multiplexed images pose unique challenges with their higher dimensionality and overlapping structures. While MaskDINO has demonstrated robust performance on natural RGB images, its adaptation for spatial omics data analysis could bridge a critical gap, enabling more accurate and efficient segmentation and classification.
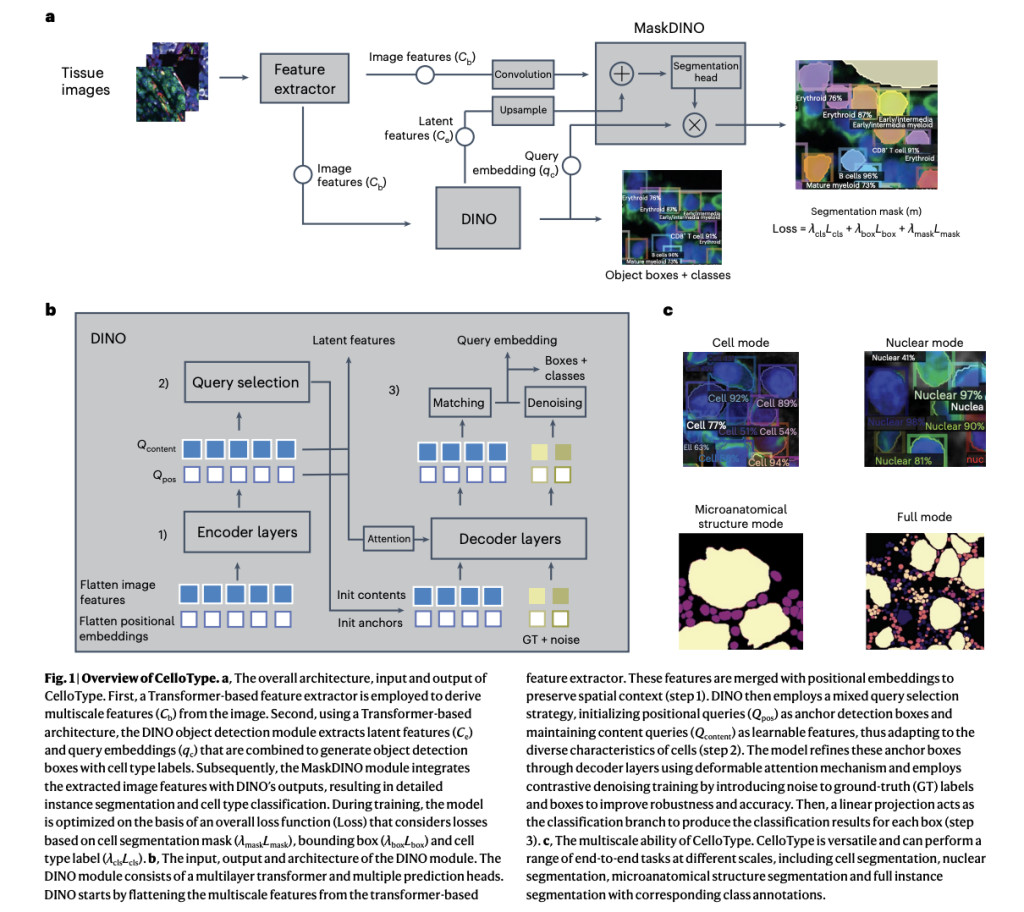
CelloType, developed by researchers from the University of Pennsylvania and the University of Iowa, is an advanced model designed to simultaneously perform cell segmentation and classification for image-based spatial omics data. Unlike conventional two-step approaches, it employs a multitask learning framework to enhance accuracy in both tasks using transformer-based architectures. The model integrates DINO and MaskDINO modules for object detection, instance segmentation, and classification, optimized through a unified loss function. CelloType also supports multiscale segmentation, enabling precise annotation of cellular and noncellular structures in tissue analysis, demonstrating superior performance on diverse datasets, including multiplexed fluorescence and spatial transcriptomic images.
CelloType comprises three key modules: (1) a Swin Transformer-based feature extraction module that generates multiscale image features for use in DINO and MaskDINO; (2) a DINO module for object detection and classification, utilizing positional and content queries, anchor box refinement, and denoising training; and (3) a MaskDINO module for instance segmentation, enhancing detection via a mask prediction branch. Training incorporates a composite loss function balancing classification, bounding box, and mask predictions. Implemented with Detectron2, CelloType leverages COCO-pretrained weights, Adam optimizer, and systematic evaluation for accuracy, supporting segmentation tasks across datasets like Xenium and MERFISH using multi-modal spatial signals.
CelloType is a deep learning framework designed for multiscale segmentation and classification of biomedical microscopy images, such as molecular, histological, and bright-field images. It uses Swin Transformer to extract multiscale features, DINO for object detection and bounding box prediction, and MaskDINO for refined segmentation. CelloType demonstrated superior performance over methods like Mesmer and Cellpose across diverse datasets, achieving higher precision, especially with its confidence-scoring variant, CelloType_C. It effectively handled segmentation tasks on multiplexed, diverse microscopy and spatial transcriptomics datasets. Additionally, it excels in simultaneous segmentation and classification, outperforming other methods on colorectal cancer CODEX data with high precision and adaptability.
In conclusion, CelloType is an end-to-end model for cell segmentation and classification in spatial omics data, combining these tasks through multitasking learning to enhance overall performance. Advanced transformer-based techniques, including Swin Transformers and the DINO module, improve object detection, segmentation, and classification accuracy. Unlike traditional methods, CelloType integrates these processes, achieving superior results on multiplexed fluorescence and spatial transcriptomic images. It also supports multiscale segmentation of cellular and non-cellular structures, demonstrating its utility for automated tissue annotation. Future improvements, including few-shot and contrastive learning, aim to address limitations in training data and challenges with spatial transcriptomics analysis.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter.. Don’t Forget to join our 55k+ ML SubReddit.
The post CelloType: A Transformer-Based AI Framework for Multitask Cell Segmentation and Classification in Spatial Omics appeared first on MarkTechPost.
Source: Read MoreÂ



 ‘
‘