Over more than three billion years, natural evolution has intricately shaped the proteins we see today. Through countless random mutations and selective pressures, nature has crafted these proteins, reflecting the deep biological principles that govern life. Modern gene sequencing unravels the immense diversity of these protein sequences and structures, revealing patterns shaped by evolutionary forces. Researchers are increasingly using large language models to decode this ‘protein language,’ discovering that these models, even without specific training on biological functions, can naturally learn to represent protein structures and functions, with their capabilities expanding significantly as they scale up in complexity and data.
Researchers from Evolutionary Scale PBC, Arc Institute, and the University of California have developed ESM3, an advanced generative language model for proteins. ESM3 can simulate evolutionary processes to create functional proteins vastly different from known ones. It integrates sequence, structure, and function to generate proteins following complex prompts. Notably, ESM3 generated a new fluorescent protein, esmGFP, which is 58% different from any known fluorescent proteins—a degree of difference comparable to 500 million years of natural evolution. This breakthrough demonstrates ESM3’s potential in protein engineering, offering creative solutions to biological challenges.
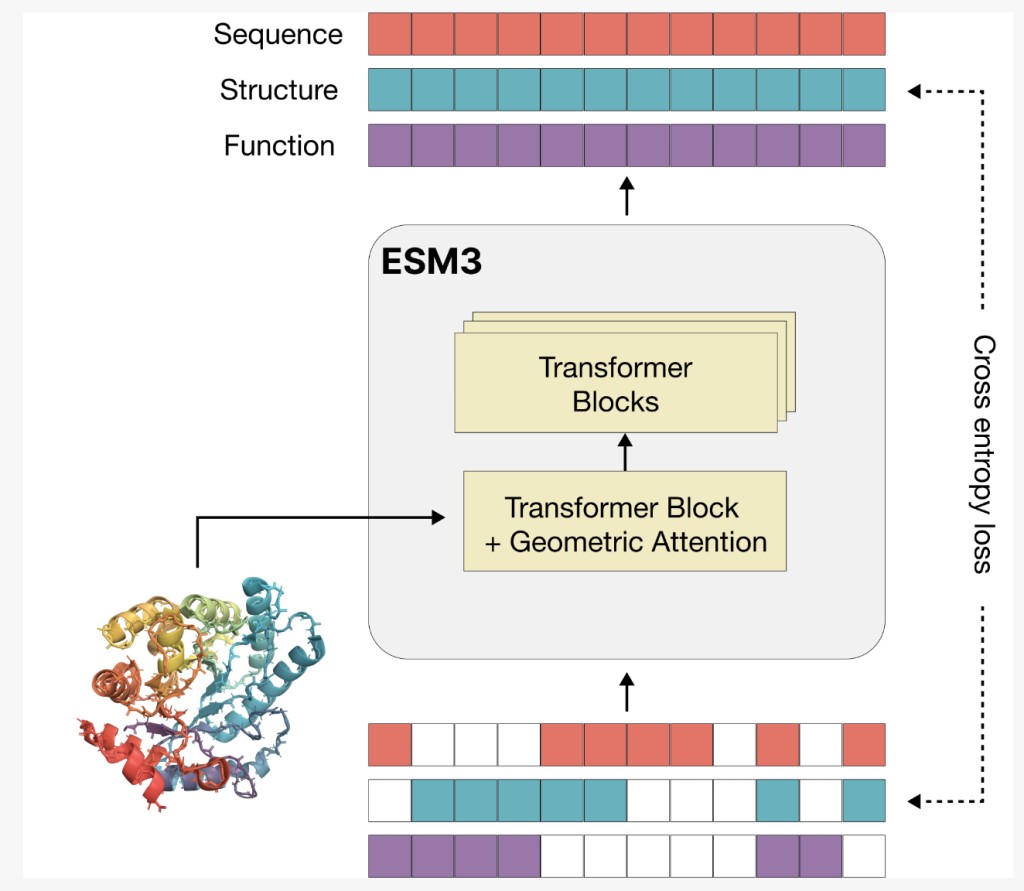
ESM3 is a sophisticated generative language model designed to understand and predict proteins’ sequence, structure, and function using tokenized data. It employs a masked language modeling approach to predict masked portions of protein data across various masking rates. ESM3 integrates sequence, structure, and function into a unified latent space and processes these modalities through transformer blocks with geometric attention. Trained on vast datasets, including 2.78 billion proteins and 236 million structures, ESM3 scales up to 98 billion parameters. Its tokenization method efficiently captures atomic details, enabling high accuracy in generating and reconstructing protein structures.
ESM3, a language model with up to 98 billion parameters, effectively predicts and generates protein sequences, structures, and functions. It processes these aspects through transformer blocks with geometric attention, training on a vast natural and synthetic protein dataset. ESM3’s generative capabilities allow it to create diverse, high-quality proteins that differ significantly from known natural proteins. It excels at following prompts from various inputs, like sequence or structural details, and can innovate within these constraints, producing novel protein designs. This versatility facilitates advanced, programmable protein design and exploration beyond natural evolutionary patterns.
Scaling and fine-tuning ESM3 models significantly enhance their ability to generate proteins that align with complex prompts, such as specific atomic coordination and structural motifs. Although the base models, trained on extensive protein datasets, perform well, fine-tuning with preference data—pairing high and low-quality outputs—reveals latent capabilities. This alignment, especially in larger models, doubles the success rate in generating accurate protein structures and increases the diversity of successful solutions. The process demonstrates that larger models have a greater inherent ability to adapt to challenging tasks, showing improved performance when aligned with specific objectives.
ESM3, a language model trained on protein sequences, generated a green fluorescent protein (GFP) with minimal similarity to existing ones. By prompting the model with critical residues and structures necessary for GFP function, ESM3 created thousands of potential designs. From these, a unique fluorescent protein, esmGFP, was identified, which differed significantly from known proteins and exhibited natural GFP-like fluorescence. This process mirrors evolutionary paths, suggesting ESM3 can explore protein spaces that evolution hasn’t, effectively simulating millions of years of evolutionary potential in generating new functional proteins.
Check out the Paper and Details. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter.Â
Join our Telegram Channel and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 45k+ ML SubReddit
Create, edit, and augment tabular data with the first compound AI system, Gretel Navigator, now generally available! [Advertisement]
The post EvolutionaryScale Introduces ESM3: A Frontier Multimodal Generative Language Model that Reasons Over the Sequence, Structure, and Function of Proteins appeared first on MarkTechPost.
Source: Read MoreÂ