Biomedical natural language processing (NLP) focuses on developing machine learning models to interpret and analyze medical texts. These models assist with diagnostics, treatment recommendations, and extracting medical information, significantly improving healthcare delivery and clinical decision-making. By processing vast amounts of biomedical literature and patient records, these models help identify patterns and insights that can lead to better patient outcomes and more informed medical research.
A significant challenge in biomedical NLP is ensuring that language models are robust and accurate when dealing with diverse and context-specific medical terminologies. Variations in drug names, particularly between generic and brand names, can cause inconsistencies and errors in model outputs. These inconsistencies can impact patient care and clinical decisions, as different terminologies may be used interchangeably by healthcare professionals and patients, potentially leading to misunderstandings or incorrect medical advice.
Existing benchmarks like MedQA and MedMCQA are commonly used to evaluate the performance of language models in the medical field. However, these benchmarks often need to account for the variability in drug nomenclature, affecting the models’ robustness and accuracy. Current models need help with synonymy and the context-specific nature of medical language, highlighting a significant gap in effective robustness evaluation. This gap points to a critical need for more specialized benchmarks to assess how well models handle real-world medical terminologies accurately.
Researchers from MIT, Harvard, and Mass General Brigham along with other prominent institutes have introduced a novel robustness evaluation method to address this issue. They developed a specialized dataset called RABBITS (Robust Assessment of Biomedical Benchmarks Involving Drug Term Substitutions) to evaluate language model performance by swapping brand and generic drug names. This innovative approach aims to simulate the real-world variability in drug nomenclature and provide a more accurate assessment of language models’ capabilities in handling medical terminology.
The RABBITS dataset was created by employing physician expert annotators to replace brand names with their generic counterparts systematically and vice versa in existing benchmarks like MedQA and MedMCQA. This transformation involved multiple rounds of expert review to ensure accuracy and contextual consistency. The process included using the RxNorm ontology to extract combinations of brand and generic drug names, resulting in 2,271 generic drugs mapped to 6,961 brands. Regular expressions identified and replaced drug names in the benchmark questions and answers. Two physicians reviewed the dataset to ensure the replacements were accurate and contextually appropriate, eliminating ambiguities and inconsistencies.
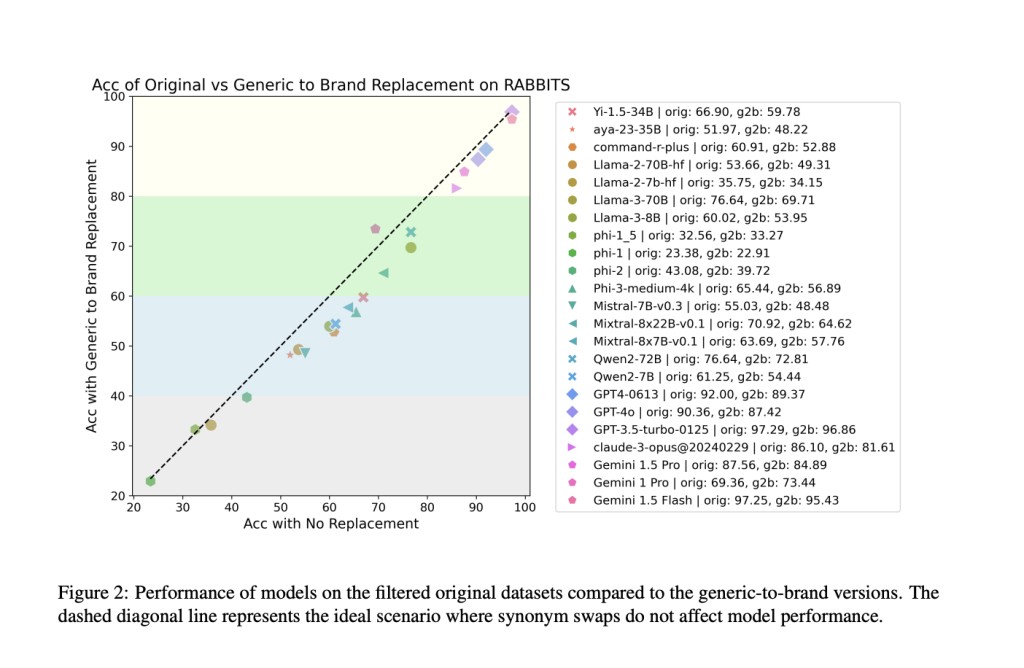
The study revealed significant performance drops in large language models (LLMs) when substituting drug names. For example, the Llama-3-70B model, which had an accuracy of 76.6% on the original dataset, dropped to 69.7% accuracy with generic-to-brand swaps. This represents a 6.9% decrease in accuracy. The researchers found that open-source models from 7B and above consistently fell below the ideal line of robustness, indicating decreased performance when drug names were swapped. The performance drop was more pronounced in the MedMCQA dataset than MedQA, with MedMCQA showing an average 4% drop in accuracy. The researchers identified dataset contamination as a potential source of this fragility, noting that pretraining datasets contained substantial benchmark test data.
To further evaluate the models, the researchers used multiple-choice questions to test the models’ ability to map brand-to-generic drug pairs and vice versa. They found that larger models, with more than 13 billion parameters, consistently outperformed smaller models in this task. However, despite this high accuracy, the models still showed significant performance drops when evaluated with the RABBITS dataset. This suggests that the models’ performance may be driven more by memorization of training data rather than genuine problem-solving skills and understanding of medical terminology.
To conclude, the research underscores a critical issue in biomedical NLP: the fragility of language models to variations in drug names. By introducing the RABBITS dataset, the research team has provided a valuable tool for assessing and improving the robustness of language models in handling medical terminology. This work reiterates the importance of developing NLP systems that are robust, context-aware, and capable of providing accurate medical information regardless of terminology variations. The development of such systems is crucial for ensuring reliable and accurate medical information processing, ultimately leading to better patient outcomes and more effective healthcare delivery.
Check out the Paper, GitHub, and Leaderboard. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter.Â
Join our Telegram Channel and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 45k+ ML SubReddit
The post RABBITS: A Specialized Dataset and Leaderboard to Aid in Evaluating LLM Performance in Healthcare appeared first on MarkTechPost.
Source: Read MoreÂ