Deep learning methods excel in detecting cardiovascular diseases from ECGs, matching or surpassing the diagnostic performance of healthcare professionals. However, due to a lack of interpretability, their “black-box†nature limits clinical adoption. Explainable AI (xAI) methods, such as saliency maps and attention mechanisms, attempt to clarify these models by highlighting key ECG features. Despite high accuracy, many models are tested on limited datasets, raising concerns about their reliability in diverse, real-world clinical scenarios. These models must provide accurate predictions and trustworthy, interpretable insights for true clinical integration.
Researchers at the Institute of Biomedical Engineering, TU Dresden, developed a deep learning architecture, xECGArch, for interpretable ECG analysis. xECGArch uniquely separates short-term (morphological) and long-term (rhythmic) ECG features using two independent Convolutional Neural Networks CNNs. The architecture was optimized for atrial fibrillation (AF) detection across four public ECG databases, achieving a 95.43% F1 score on unseen data. Deep Taylor decomposition was identified as the most trustworthy xAI method among the 13 tested using perturbation analysis. This approach enhances the interpretability and reliability of ECG classifications, bridging the gap between clinical needs and automated analysis.
The study utilized four extensive 12-lead ECG databases: PTB-XL, Georgia-12-Lead, China Physiological Signal Challenge 2018 (CPSC2018), and Chapman-Shaoxing, all sampled at 500 Hz. Given the emphasis on single-lead ECGs, only lead II was used to ensure applicability for portable devices and effectiveness in detecting AF. The datasets, containing both AF and non-AF recordings, were balanced to include 4,927 samples from each class, addressing the issue of classifier bias towards more prevalent courses. ECG signals were preprocessed through high-pass filtering, noise reduction using discrete wavelet transformation, and scaling.Â
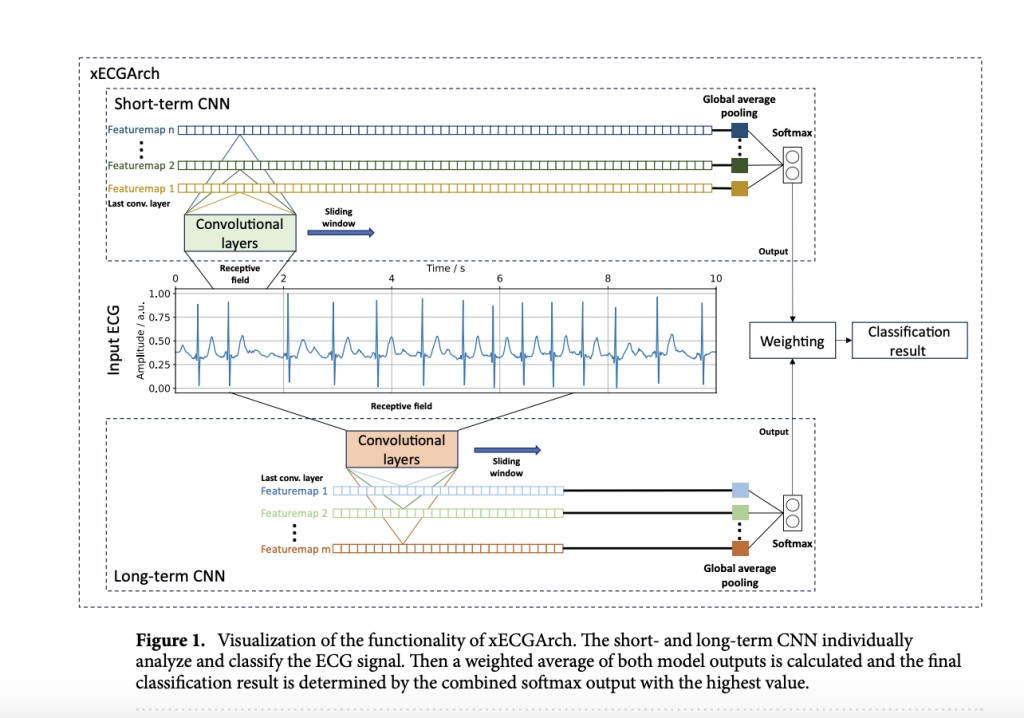
The xECGArch deep learning architecture designed for ECG analysis integrates two independent 1D CNNs focusing on short-term and long-term ECG features, which are crucial for interpreting morphological and rhythmic patterns. The short-term network analyzes rapid, beat-level features with a receptive field of 0.6 seconds, while the long-term network covers the entire 10-second ECG recording to capture broader rhythmic information. Both networks employ global average pooling (GAP) to reduce input dimensions before classification via a softmax layer, enhancing efficiency and performance. To ensure robustness and interpretability, xECGArch underwent extensive hyperparameter tuning and cross-validation. Various xAI methods were employed and evaluated for interpretability, including gradient-based techniques, decomposition methods like deep Taylor decomposition (DTD) and Layer-wise Relevance Propagation (LRP), GradCAM variants, and SHAP values. These methods offer insights into the model’s decision-making by highlighting relevant features and contributions within the ECG data.
The xECGArch, a deep learning framework, was designed to classify AF in 10-second ECG recordings. It comprises short-term and long-term CNNs tailored to capture different temporal features. The short-term CNN focuses on a 0.6-second window, suitable for individual heartbeats, while the long-term CNN covers the entire 10-second recording. The best short-term model achieved a 94.18% F1 score, while the best long-term model reached 95.13%. Combining their outputs via weighted averaging improved the overall performance to a 95.43% F1 score. DTD and other methods like Integrated Gradients (ITG) and LRP were used for model interpretation, revealing that the short-term model emphasized P waves and F waves, while the long-term model focused on irregular R peaks. This multi-scale approach enhances accuracy and interpretability in AF detection from ECG signals.
In conclusion, The xECGArch’s combined short- and long-term CNNs enhance AF detection by leveraging distinct temporal features. The model surpasses many existing methods, achieving a high F1 score of 95.43%, although some reported higher scores on less diverse datasets. Explanation methods like DTD proved effective for interpreting model decisions, highlighting relevant ECG features such as P waves for non-AF and irregular QRS complexes for AF. This multi-scale approach improves diagnostic accuracy and enhances the interpretability of ECG analysis. Future applications include other biosignals and improving big data cardiac screening through automated, trustworthy diagnostics.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our Telegram Channel, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 44k+ ML SubReddit
The post xECGArch: A Multi-Scale Convolutional Neural Network CNN for Accurate and Interpretable Atrial Fibrillation Detection in ECG Analysis appeared first on MarkTechPost.
Source: Read MoreÂ